The intracellular virus-host interface of henipaviruses
- PMID: 40679287
- PMCID: PMC12363173
- DOI: 10.1128/jvi.00770-25
The intracellular virus-host interface of henipaviruses
Abstract
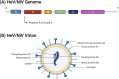
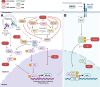
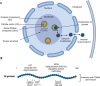
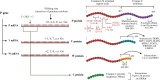
The Henipavirus genus comprises five viral species, of which the prototype members, Hendra virus (HeV) and Nipah virus (NiV), are reported to infect humans. In humans and other spill-over hosts, HeV/NiV can cause severe respiratory and/or encephalitic disease, with mortality rates exceeding 50%; currently, there are no approved human vaccines and only limited therapeutic options. As members of the family Paramyxoviridae, henipaviruses have six "core" structural proteins and typically three additional accessory proteins that are expressed from the P gene. Several of these proteins are multifunctional, with roles in forming intracellular interfaces with the host (in particular, M, P, V, W, and C proteins), to modulate processes including antiviral responses, supporting viral replication. Understanding the molecular basis of these interfaces and their functions is critical to delineate the mechanisms of pathogenesis and may inform new strategies to combat infection and disease. Recent research has significantly advanced the understanding of the functions and interactions of multifunctional intracellular henipavirus proteins, including revealing novel roles in subverting the nucleolar DNA damage response (DDR) and modulating the functions of 14-3-3 proteins. This review will discuss the intracellular virus-host interface, focusing on the M, P, V, W, and C proteins of HeV/NiV, with a focus on recently identified functions and interactions.
Keywords: Hendra virus; Nipah virus; henipavirus; immune evasion; viral proteins; virus-host interface.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- ICTV . 2025. ICTV genus: Henipavirus. Available from: https://ictv.global/report/chapter/paramyxoviridae/paramyxoviridae/henip...
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

