Potential in-host evolution of Klebsiella pneumoniae ST147: convergence and the role of capsular alterations in morphotype diversity
- PMID: 40679293
- PMCID: PMC12403872
- DOI: 10.1128/spectrum.00170-25
Potential in-host evolution of Klebsiella pneumoniae ST147: convergence and the role of capsular alterations in morphotype diversity
Abstract
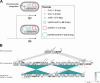
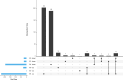
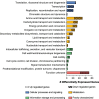
Klebsiella pneumoniae, an important opportunistic pathogen, is traditionally classified into classic and hypervirulent pathotypes. Convergent strains combining antimicrobial resistance (AMR) and hypervirulence have emerged globally, posing a significant health challenge. In this study, we investigated potential in-host evolution and morphotypic variation among consecutive K. pneumoniae ST147 isolates from a single patient. As described in a previous study, the isolates displayed distinct morphotypes-small white, normal white, gray, and gray/dry (g/d) colonies. In this study, we now present new genomic and phenotypic analyses, which suggest the acquisition of AMR and virulence genes through plasmid gain. The early isolates carried fewer plasmids, resulting in low resistance and virulence, while later isolates acquired a hybrid IncFIB-IncHI1B plasmid encoding different resistance and aerobactin genes, significantly increasing their geno- and phenotypic AMR and virulence potential. Chromosomal integration of this plasmid in one isolate stabilized the acquired traits. Disruptions in the K locus, mediated by insertion sequences, were linked to the gray and g/d morphotypes, impairing capsule biosynthesis. Uronic acid assays confirmed reduced capsule production in these isolates. In contrast, small colony variants showed significant transcriptomic changes, including upregulation of capsule biosynthesis, iron uptake pathways, and AMR genes, suggesting persistence through altered metabolism. Our findings suggest in-host microevolution of K. pneumoniae ST147 from a classic to a convergent pathotype and highlight the genomic and transcriptomic adaptations underlying morphotypic diversity, providing new insights into the pathogen's adaptability and persistence in certain environments.IMPORTANCEKlebsiella pneumoniae is an important bacterial pathogen that can cause severe infections, particularly in healthcare settings. Traditionally, strains are classified as either drug-resistant or highly virulent, but recent strains combining both features have led to hard-to-treat infections. In this study, we investigated the potential in-host evolution of K. pneumoniae ST147 isolates from a single patient, revealing key genomic and phenotypic changes driving their adaptability. Over time, isolates acquired additional plasmids carrying antimicrobial resistance and virulence genes, including a hybrid plasmid integrated into the bacterial chromosome, stabilizing its beneficial traits. In addition, morphotypic diversity emerged, linked to genomic disruptions in capsule biosynthesis genes (K locus). While capsule-deficient morphotypes displayed structural changes, small colony variants exhibited transcriptomic adaptations to persist under stress. This work underscores the dynamic evolutionary capacity of K. pneumoniae adapting within the host, providing critical insights into its persistence, resilience, and the challenges posed by emerging convergent strains in clinical settings.
Keywords: K. pneumoniae; SCV; capsule deficiencies; convergent pathotype; in-host evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Heiden SE, Hübner N-O, Bohnert JA, Heidecke C-D, Kramer A, Balau V, Gierer W, Schaefer S, Eckmanns T, Gatermann S, Eger E, Guenther S, Becker K, Schaufler K. 2020. A Klebsiella pneumoniae ST307 outbreak clone from Germany demonstrates features of extensive drug resistance, hypermucoviscosity, and enhanced iron acquisition. Genome Med 12:113. doi: 10.1186/s13073-020-00814-6 - DOI - PMC - PubMed
-
- Li P, Liang Q, Liu W, Zheng B, Liu L, Wang W, Xu Z, Huang M, Feng Y. 2021. Convergence of carbapenem resistance and hypervirulence in a highly-transmissible ST11 clone of K. pneumoniae: an epidemiological, genomic and functional study. Virulence 12:377–388. doi: 10.1080/21505594.2020.1867468 - DOI - PMC - PubMed
-
- Shaidullina ER, Schwabe M, Rohde T, Shapovalova VV, Dyachkova MS, Matsvay AD, Savochkina YA, Shelenkov AA, Mikhaylova YV, Sydow K. 2023. Genomic analysis of the international high-risk clonal lineage Klebsiella pneumoniae sequence type 395. Genome Med 15:9. doi: 10.1186/s13073-023-01159-6 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

