Engineering Streptomyces coelicolor for heterologous expression of the thiopeptide GE2270A-A cautionary tale
- PMID: 40679465
- PMCID: PMC12309360
- DOI: 10.1093/jimb/kuaf019
Engineering Streptomyces coelicolor for heterologous expression of the thiopeptide GE2270A-A cautionary tale
Abstract
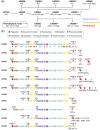
The thiopeptide GE2270A is a clinically relevant, ribosomally synthesised, and post-translationally modified peptide naturally produced by Planobispora rosea. Due to the genetically intractable nature of P. rosea, heterologous expression is considered a possible route to yield improvement. In this study, we focused on improving GE2270A production through heterologous expression of the biosynthetic gene cluster (BGC) in the model organism Streptomyces coelicolor M1146. A statistically significant yield improvement was obtained in the S. coelicolor system through the data-driven rational engineering of the BGC, including the introduction of additional copies of key biosynthetic and regulatory genes. However, despite our best efforts, the highest production level observed in the strains generated in this study is 12× lower than published titres achieved in the natural producer and 50× lower than published titres obtained using Nonomuraea ATCC 39727 as expression host. These results suggest that, while using the most genetically amenable strain as host can be the right choice when exploring different BGC designs, the choice of the most suitable host has a major effect on the achievable yield and should be carefully considered. The analysis of the multiomics data obtained in this study suggests an important role of PbtX in GE2270A biosynthesis and provides insights into the differences in production metabolic profiles between the different strains. One Sentence Summary: Data-driven rational engineering of Streptomyces coelicolor for heterologous production of the thiopeptide antibiotic GE2270A resulted in increased production but encountered unexpected challenges compared to production in the natural producer or the alternative host Nonomuraea ATCC 39727.
Keywords: Streptomyces coelicolor; GE2270A; Metabolic engineering; Synthetic biology; Thiopeptide.
© The Author(s) 2025. Published by Oxford University Press on behalf of Society of Industrial Microbiology and Biotechnology.
Conflict of interest statement
There are no conflicts of interest.
Figures


References
-
- Abramson J., Adler J., Dunger J., Evans R., Green T., Pritzel A., Ronneberger O., Willmore L., Ballard A. J., Bambrick J., Bodenstein S. W., Evans D. A., Hung C.-C., O'Neill M., Reiman D., Tunyasuvunakool K., Wu Z., Žemgulytė A., Arvaniti E., Jumper J. M. (2024). Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature, 630(8016), 493–500. 10.1038/s41586-024-07487-w - DOI - PMC - PubMed
-
- Arnison P. G., Bibb M. J., Bierbaum G., Bowers A. A., Bugni T. S., Bulaj G., Camarero J. A., Campopiano D. J., Challis G. L., Clardy J., Cotter P. D., Craik D. J., Dawson M., Dittmann E., Donadio S., Dorrestein P. C., Entian K.-D., Fischbach M. A., Garavelli J. S., van der Donk W. A. (2013). Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Natural Product Reports, 30(1), 108–160. 10.1039/C2NP20085F - DOI - PMC - PubMed
-
- Bai C., Zhang Y., Zhao X., Hu Y., Xiang S., Miao J., Lou C., Zhang L. (2015). Exploiting a precise design of universal synthetic modular regulatory elements to unlock the microbial natural products in Streptomyces. Proceedings of the National Academy of Sciences, 112(39), 12181–12186. 10.1073/pnas.1511027112 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

