Endogenous retrovirus-like proteins recruit UBQLN2 to stress granules and shape their functional biology
- PMID: 40680123
- PMCID: PMC12273755
- DOI: 10.1126/sciadv.adu6354
Endogenous retrovirus-like proteins recruit UBQLN2 to stress granules and shape their functional biology
Abstract
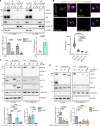
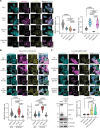
The human genome is replete with sequences derived from foreign elements including endogenous retrovirus-like proteins of unknown function. Here, we show that UBQLN2, a ubiquitin-proteasome shuttle factor implicated in neurodegenerative diseases, is regulated by the linked actions of two retrovirus-like proteins, retrotransposon gag-like 8 (RTL8) and paternally expressed gene 10 (PEG10). RTL8 confers on UBQLN2 the ability to complex with and regulate PEG10. PEG10, a core component of stress granules, drives the recruitment of UBQLN2 to stress granules under various stress conditions but can only do so when RTL8 is present. Changes in UBQLN2, RTL8, or PEG10 levels further remodel the kinetics of stress granule disassembly and translation recovery. PEG10 also alters overall stress granule composition by incorporating select extracellular vesicle proteins. Within stress granules, PEG10 forms virus-like particles, underscoring the structural heterogeneity of this class of biomolecular condensates. Together, these results reveal an unexpected link between pathways of cellular proteostasis and endogenous retrovirus-like proteins.
Figures





Update of
-
Endogenous retrovirus-like proteins recruit UBQLN2 to stress granules and alter their functional properties.bioRxiv [Preprint]. 2024 Oct 25:2024.10.24.620053. doi: 10.1101/2024.10.24.620053. bioRxiv. 2024. Update in: Sci Adv. 2025 Jul 18;11(29):eadu6354. doi: 10.1126/sciadv.adu6354. PMID: 39484508 Free PMC article. Updated. Preprint.
References
-
- Pastuzyn E. D., Day C. E., Kearns R. B., Kyrke-Smith M., Taibi A. V., McCormick J., Yoder N., Belnap D. M., Erlendsson S., Morado D. R., Briggs J. A. G., Feschotte C., Shepherd J. D., The neuronal gene Arc encodes a repurposed retrotransposon Gag protein that mediates intercellular RNA transfer. Cell 173, 275 (2018). - PMC - PubMed
-
- Pang S. W., Lahiri C., Poh C. L., Tan K. O., PNMA family: Protein interaction network and cell signalling pathways implicated in cancer and apoptosis. Cell. Signal. 45, 54–62 (2018). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

