Role of d-serine in intestinal ROS accumulation after sleep deprivation
- PMID: 40680136
- PMCID: PMC12273795
- DOI: 10.1126/sciadv.adr8592
Role of d-serine in intestinal ROS accumulation after sleep deprivation
Abstract
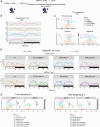
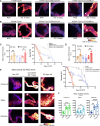
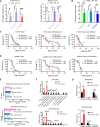
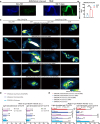
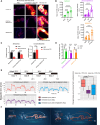
Prolonged sleep deprivation (SD) results in increased accumulation of reactive oxygen species (ROS) in gut, although the underlying mechanisms remain to be elucidated. This study identifies d-serine as a crucial regulator of gut ROS during SD. Knockdown of serine racemase (SR), the enzyme responsible for d-serine production, prevents the enhanced ROS buildup during SD in Drosophila. Gut enterocytes (ECs) respond to γ-aminobutyric acid (GABA) signaling by producing d-serine, which influences downstream N-methyl-d-aspartate receptor (NMDAR) activity and modulates sleep pressure. However, the continuous demand for sleep disrupts this feedback loop. Prolonged SD leads to increased levels of d-serine in the gut, an elevated pyruvate pool in ECs, enhanced mitochondrial oxidative phosphorylation, impaired lipid metabolism in peroxisomes, and the accumulation of harmful ROS. In conclusion, our findings illuminate the metabolic alterations and brain-gut communication pathways that may contribute to the increase in gut d-serine and subsequent ROS accumulation induced by SD.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

