Single-cell transcriptomics reveal individual and cooperative effects of trisomy 21 and GATA1s on hematopoiesis
- PMID: 40680731
- PMCID: PMC12365824
- DOI: 10.1016/j.stemcr.2025.102577
Single-cell transcriptomics reveal individual and cooperative effects of trisomy 21 and GATA1s on hematopoiesis
Abstract
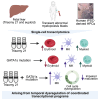
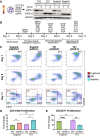
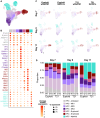
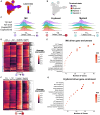
Trisomy 21 (T21) is associated with baseline erythrocytosis, thrombocytopenia, neutrophilia, transient abnormal myelopoiesis (TAM), and myeloid leukemia of Down syndrome (ML-DS). TAM and ML-DS blasts harbor mutations in GATA1, resulting in the exclusive expression of the truncated isoform GATA1s. Germline GATA1s mutations in individuals without T21 cause congenital cytopenias, typically without a leukemic predisposition. To dissect the developmental effects of T21 and GATA1s, we used a combination of isogenic human induced pluripotent stem cells, primary human fetal and neonatal cells, and single-cell transcriptomics to interrogate hematopoietic progenitors differing only by chromosome 21 and/or GATA1 status. Both T21 and GATA1s induced early lineage skewing, and trajectory analysis revealed that GATA1s altered the temporal regulation of lineage-specific transcriptional programs, disrupting cell proliferation and maturation irrespective of chromosomal context. These studies uncovered unexpected heterogeneity and lineage priming in early, multipotent hematopoietic progenitors and identified transcriptional and functional maturation blocks linked to GATA1s.
Keywords: Down syndrome; GATA1; GATA1s; Trisomy 21; hematopoiesis; hematopoietic progenitors; iPSC; lineage priming; single-cell RNA sequencing; transient abnormal myelopoiesis.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


Update of
-
Single-cell transcriptomics reveal individual and synergistic effects of Trisomy 21 and GATA1s on hematopoiesis.bioRxiv [Preprint]. 2024 Oct 31:2024.05.24.595827. doi: 10.1101/2024.05.24.595827. bioRxiv. 2024. Update in: Stem Cell Reports. 2025 Aug 12;20(8):102577. doi: 10.1016/j.stemcr.2025.102577. PMID: 38826323 Free PMC article. Updated. Preprint.
References
-
- Alford K.A., Reinhardt K., Garnett C., Norton A., Böhmer K., Von Neuhoff C., Kolenova A., Marchi E., Klusmann J.H., Roberts I., et al. Analysis of GATA1 mutations in down syndrome transient myeloproliferative disorder and myeloid leukemia. Blood. 2011;118:2222–2238. doi: 10.1182/blood-2011-03-342774. - DOI - PubMed
-
- Banno K., Omori S., Hirata K., Nawa N., Nakagawa N., Nishimura K., Ohtaka M., Nakanishi M., Sakuma T., Yamamoto T., et al. Systematic Cellular Disease Models Reveal Synergistic Interaction of Trisomy 21 and GATA1 Mutations in Hematopoietic Abnormalities. Cell Rep. 2016;15:1228–1241. doi: 10.1016/j.celrep.2016.04.031. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

