Targeting Hepatic Stellate Cell PD-L1 Alters Liver Inflammation and Fibrosis in CCl4 Liver Injury Mouse Model
- PMID: 40681040
- PMCID: PMC12419031
- DOI: 10.1016/j.jcmgh.2025.101587
Targeting Hepatic Stellate Cell PD-L1 Alters Liver Inflammation and Fibrosis in CCl4 Liver Injury Mouse Model
Abstract
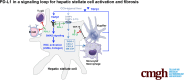
Background & aims: Programmed death-ligand 1 (PD-L1) on hepatic stellate cells (HSCs) is required for HSC activation and suppressing T and B lymphocytes. We tested whether targeting HSC PD-L1 influenced liver inflammation and fibrosis in a carbon tetrachloride (CCl4) injury mouse model.
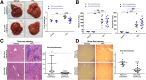
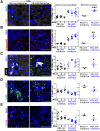
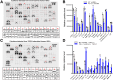
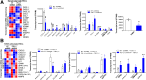
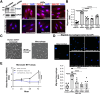
Methods: HSC-specific PD-L1 knockout (PD-L1HSCKO) mice were created by crossing Cd274 floxed mice to Collagen1A1-Cre mice. CCl4 was injected into PD-L1HSCKO and PD-L1HSCWT mice twice weekly for 6 weeks. Liver fibrosis was assessed by Trichrome and Picrosirius Red staining; HSC activation was determined by immunofluorescence and Western blot for HSC activation markers; liver inflammation was studied by multiplex immunofluorescence and cytokine profiling. Multiomics was leveraged to determine how targeting PD-L1 altered HSC producing collagens and cytokines/chemokines.
Results: Collagen deposition was reduced in CCl4-injured PD-L1HSCKO livers compared with CCl4-injured PD-L1HSCWT livers; myofibroblast density was lower in CCl4-injured PD-L1HSCKO livers compared with CCl4-injured PD-L1HSCWT livers. CCl4-injured PD-L1HSCKO livers had higher lymphocyte densities (GranzymeB+, CD8a+, CD20+) but lower Kupffer and myeloid cell densities (F4/80+ and CD11b+) compared with CCl4-injured PD-L1HSCWT livers. Serum aspartate aminotransferase and alanine aminotransferase, however, were similarly elevated by CCl4 in both groups. Spatial and bulk-cell transcriptomics revealed a global transcriptomic change of HSCs induced by PD-L1 targeting. A targeted proteomics identified that HSC secretion of a group of cytokines/chemokines, including growth/differentiation factor 15, granulocyte-macrophage colony-stimulating factor, C-X-C motif and C-C motif chemokines, was altered upon PD-L1 targeting, highlighting the role of HSC PD-L1 in HSC/Kupffer and HSC/myeloid cell interactions during HSC activation and fibrosis development.
Conclusions: Targeting HSC PD-L1 altered HSC transcriptome and liver inflammation, and suppressed liver fibrosis, representing a potential therapeutic strategy for liver fibrosis.
Keywords: Gene Set Enrichment Analysis; Glial Fibrillary Acidic Protein (GFAP); Macrophage; Single-cell RNA Sequencing; Transforming Growth Factor Beta.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Figures







References
-
- Hammerich L., Tacke F. Hepatic inflammatory responses in liver fibrosis. Nat Rev Gastroenterol Hepatol. 2023;20:633–646. - PubMed
-
- Kisseleva T., Brenner D. Molecular and cellular mechanisms of liver fibrosis and its regression. Nat Rev Gastroenterol Hepatol. 2021;18:151–166. - PubMed
-
- Tsuchida T., Friedman S.L. Mechanisms of hepatic stellate cell activation. Nat Rev Gastroenterol Hepatol. 2017;14:397–411. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

