GPD2 inhibition impairs coagulation function via ROS/NF-κB/P2Y12 pathway
- PMID: 40682019
- PMCID: PMC12273321
- DOI: 10.1186/s11658-025-00759-x
GPD2 inhibition impairs coagulation function via ROS/NF-κB/P2Y12 pathway
Abstract
Background: Coronary heart disease (CHD) remains a global health threat. As antiplatelet therapy constitutes the cornerstone of CHD management, ticagrelor has been universally endorsed as a first-line agent in major clinical guidelines. However, the therapeutic efficacy of ticagrelor is compromised by interindividual variability in bleeding risk. Notably, while inherited genetic variations account for part of this heterogeneity, the dynamic regulatory role of modifiable epigenetic mechanisms-particularly DNA methylation in mediating platelet reactivity-remains inadequately characterized, presenting a critical knowledge gap in optimizing precision antiplatelet strategies.
Methods: We utilized the 850k methylation array to measure DNA methylation levels in blood samples from 47 healthy controls and 93 patients with CHD. Subsequently, epigenome-wide association study (EWAS), summary data-based Mendelian randomization (SMR), and heterogeneity in dependent instruments (HEIDI) analyses were applied to pinpoint critical methylation sites that influence gene expression, platelet function recovery, and bleeding risk. After developing a targeted cellular model using the CRISPR-dCas9-DNMT3A/Tet1CD-U6-sgRNA system and integrating with transcriptomic sequencing data, we conducted mechanistic cellular experiments to elucidate how these methylation sites affect platelet function recovery and bleeding risk. The findings were further validated through animal studies.
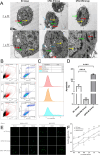
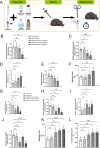
Results: Integrated analysis of EWAS and SMR-HEIDI revealed that hypermethylation at CpG site cg03230175 within the GPD2 gene promoter region was significantly associated with decreased GPD2 gene expression (P = 1.76E-18), delayed platelet functional recovery (P = 9.02 × 10-3), and elevated hemorrhagic risk (P = 2.71 × 10-2). Transcriptomic studies indicated that GPD2 gene (cg03230175) methylation affects mitochondrial function, nuclear factor kappa B (NF-κB) signaling pathway, reactive oxygen species metabolic process, and G protein-coupled receptor (GPCR) ligand binding. Cellular experiments demonstrated that the GPD2 gene (cg03230175) methylation inhibits coagulation function by suppressing reactive oxygen species (ROS) production, NF-κB activation, and P2Y12 gene expression (P2Y12 receptor plays a pivotal role in platelet activation, thrombus formation, and the pathogenesis of thrombotic disorders). The animal study results confirmed that GPD2 enzyme inhibition can indeed prolong the clotting time in mice.
Conclusions: GPD2 gene (cg03230175) methylation resulted in reduced gene expression levels, inhibited mitochondrial energy metabolism, decreased ROS levels, and affected P2Y12 gene expression through the NF-κB pathway, ultimately leading to inhibition of coagulation function. Registry: The Impact of Genotype on Pharmacokinetics and Antiplatelet Effects of Ticagrelor in Healthy Chinese (IGPPT).
Trial registration number: NCT03092076. Date of Registration: 09 March 2017, retrospectively registered. URL of trial registry record: https://clinicaltrials.gov/ct2/show/NCT03092076 .
Keywords: DNA methylation; Epigenetic mechanism; GPD2; NF-κB; P2Y12; ROS.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The Ethics Review Committee of Guangdong Provincial People’s Hospital approved our human and animal studies (no. GDREC:KY2023-090-02) on 10 March 2023, in compliance with the “Ethical Review Measures for Biomedical Research Involving Human Subjects” and “Guidance on the Humane Treatment of Laboratory Animals” issued by the Ministry of Science and Technology of China, as well as relevant international animal welfare guidelines and regulations (including the ICLAS statement). Our research strictly adhered to the Declaration of Helsinki and other applicable guidelines. The study was registered with ClinicalTrials.gov (NCT03092076) and was strictly conducted in accordance with the relevant provisions of the Declaration of Helsinki and received approval from the Medical Research Ethics Committee of Guangdong Provincial People’s Hospital (no. GDREC2015143H(R1) and no. GDREC2017071H). Each patient was fully informed of the details of the research and the entire process. Furthermore, written informed consent was obtained from all participants (no. 20150115 and no.20170211). Competing interests: The authors declare that they have no competing interests.
Figures





References
-
- Levine GN, Bates ER, Bittl JA, Brindis RG, Fihn SD, Fleisher LA, Granger CB, Lange RA, Mack MJ, Mauri L, Mehran R, Mukherjee D, Newby LK, O’Gara PT, Sabatine MS, Smith PK, Smith SC Jr. 2016 ACC/AHA guideline focused update on duration of dual antiplatelet therapy in patients with coronary artery disease: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J Am Coll Cardiol. 2016;68(10):1082–115. 10.1016/j.jacc.2016.03.513. - PubMed
-
- Corbett SJ, Ftouh S, Lewis S, Lovibond K, Guideline Committee. Acute coronary syndromes: summary of updated NICE guidance. BMJ. 2021;372:m4760. 10.1136/bmj.m4760. - PubMed
-
- Kamran H, Jneid H, Kayani WT, Virani SS, Levine GN, Nambi V, Khalid U. Oral antiplatelet therapy after acute coronary syndrome: a review. JAMA. 2021;325(15):1545-1555. 10.1001/jama.2021.0716. Erratum in: JAMA. 2021 Jul 13;326(2):190. 10.1001/jama.2021.9484. - PubMed
-
- Wallentin L, Becker RC, Budaj A, Cannon CP, Emanuelsson H, Held C, Horrow J, Husted S, James S, Katus H, Mahaffey KW, Scirica BM, Skene A, Steg PG, Storey RF, Harrington RA, Freij A, Thorsén M, PLATO Investigators. Ticagrelor versus clopidogrel in patients with acute coronary syndromes. N Engl J Med. 2009;361(11):1045–57. 10.1056/NEJMoa0904327. - PubMed
-
- Valgimigli M, Bueno H, Byrne RA, Collet JP, Costa F, Jeppsson A, Jüni P, Kastrati A, Kolh P, Mauri L, Montalescot G, Neumann FJ, Petricevic M, Roffi M, Steg PG, Windecker S, Zamorano JL, ESC Scientific Document Group, ESC Committee for Practice Guidelines (CPG), ESC National Cardiac Societies. 2017 ESC focused update on dual antiplatelet therapy in coronary artery disease developed in collaboration with EACTS: the task force for dual antiplatelet therapy in coronary artery disease of the European Society of Cardiology (ESC) and of the European Association for Cardio-Thoracic Surgery (EACTS). Eur Heart J. 2018;39(3):213–60. 10.1093/eurheartj/ehx419. - PubMed
MeSH terms
Substances
Associated data
Grants and funding
- No. 82274016/National Natural Science Foundation of China
- 81872934/National Natural Science Foundation of China
- 82003746/National Natural Science Foundation of China
- 82473910/National Natural Science Foundation of China
- 0720240120/Leading Talent of Guangdong Special Support Program
- 2021A1515220031/Guangdong Basic and Applied Basic Research Foundation
- 2023A1515011133/Basic and Applied Basic Research Foundation of Guangdong Province
- 2025A1515012395/Basic and Applied Basic Research Foundation of Guangdong Province
- No. 2023B03J1251/Science and Technology Program of Guangzhou
- No. 2019B020229003/Key-Area Research and Development Program of Guangdong Province, China
- No. 2017YFC0909301/National key research and development program
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

