Association of specific microbiota taxa in the amniotic fluid at birth with severe acute and longer-term outcomes of very preterm infants: a prospective observational study
- PMID: 40682023
- PMCID: PMC12275360
- DOI: 10.1186/s12916-025-04259-9
Association of specific microbiota taxa in the amniotic fluid at birth with severe acute and longer-term outcomes of very preterm infants: a prospective observational study
Abstract
Background: Dysbiotic microbial colonization predisposes to severe outcomes of prematurity, including mortality and severe morbidities like necrotizing enterocolitis (NEC), late-onset infection (LOI) and bronchopulmonary dysplasia (BPD). Here, we studied the variations in the bacterial signatures in the amniotic fluid (AF) of very preterm deliveries < 32 weeks with severe acute and longer-term outcomes within a prospective cohort study.
Methods: One hundred twenty-six AF samples were available for 16S rRNA gene metabarcoding to describe bacterial community structure and diversity in connection to intraventricular haemorrhage (IVH), LOI, focal intestinal perforation (FIP), NEC, retinopathy of prematurity (ROP) and the 2-year cognitive (MDI) and motor (PDI) outcome.
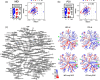
Results: Diversity and overall bacterial community composition did not differ between the studied outcomes. But disparities in sequences assigned to single bacterial taxa were observed for the acute outcomes LOI and ROP and the longer-term impairments of MDI and PDI. Enrichments associated with a poor acute outcome were particularly detected in the Escherichia-Shigella cluster, while the predominance of Ureaplasma and Enterococcus species was associated with unrestricted acute and longer-term outcomes. Analysis for FIP did not reach any significance. IVH and NEC constituted rare events, prohibiting the analyses.
Conclusions: Our data provide evidence that microbiota patterns at birth might allow the early identification of infants at risk for the severe outcomes of prematurity and argue against morbidity-specific associations. The data support the early origins hypothesis and relevant contribution of prenatal factors. The partly existing disparities between acute and longer-term outcomes might be traced back to the relevant impact of the diverse longitudinal exposures and socioeconomic factors.
Keywords: 16S rRNA; Amniotic fluid; Bronchopulmonary dysplasia; Intraventricular haemorrhage; Late-onset infection; Microbiome; Preterm infant; Psychomotor outcome; Respiratory distress; Retinopathy of prematurity.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study was conducted following the rules of the Declaration of Helsinki, was approved by the ethics committee of the Justus-Liebig-University of Gießen (Az 135/12) and registered at DRKS (DRKS00004600). Written informed consent was obtained from the parents of preterm infants and pregnant women intended for prenatal interventions after provision of oral and written information. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures


References
-
- Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. 2016;375:2369–79. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

