MIR4726EccDNA drives bortezomib resistance in multiple myeloma by enhancing MIR4726-5p/NXF1/NKIRAS2 axis dependent autophagy
- PMID: 40682103
- PMCID: PMC12273429
- DOI: 10.1186/s12964-025-02340-7
MIR4726EccDNA drives bortezomib resistance in multiple myeloma by enhancing MIR4726-5p/NXF1/NKIRAS2 axis dependent autophagy
Abstract
Background: Despite many new drugs, multiple myeloma (MM) remains an incurable plasma cell malignancy, and drug resistance is a long-standing topic in this field. Characterized by efficient transcription without being limited by the double helix structure and promoter, extrachromosomal circular DNA (EccDNA) has been proven to be widely involved in cancer development and drug resistance.
Methods: We performed circle-seq and mRNA-seq on samples from three MM patients at the time of complete response and relapse to screen EccDNA candidate molecules. Outward PCR and Sanger sequencing were used to identify EccDNA molecules. RT‒qPCR and WB were performed to detect gene expression levels. Fluorescence in situ hybridization (FISH) was carried out to detect the deletion of chromosome 17p (del (17p)). Transmission electron microscopy (TEM) was conducted to observe autophagosomes. Luciferase reporter assays were performed to validate the binding of microRNAs to target genes. Cell viability assays and apoptosis assays were employed to assess drug resistance. Xenograft tumor mouse models were established for in vivo experiments. Immunohistochemistry (IHC) was used to detect protein expression levels.
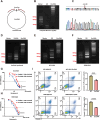
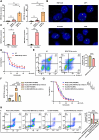
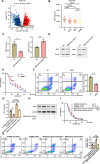
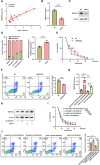
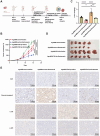
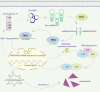
Results: We successfully identified an EccDNA molecule (EccDNAchr17:38719676-38719812) in one relapsed MM patient with del(17p) and named it MIR4726EccDNA. We demonstrated that the overexpression of MIR4726EccDNA in MM cells can increase bortezomib resistance. We further confirmed that the precursor miRNA carried by MIR4726EccDNA can be efficiently transcribed in MM cells and that MIR4726EccDNA drives bortezomib resistance via the MIR4726-5p/NXF1/NKIRAS2 axis. We further revealed that downregulation of NFKB inhibitor interacting Ras like 2 (NKIRAS2) activated the NF-κB pathway and increased autophagy. Moreover, we established a xenograft model of human MM via subcutaneous inoculation. We administered intra-tumoral injection of AgoMIR4726-5p and intraperitoneal injection of bortezomib and found that AgoMIR4726-5p promoted tumor progression and partially drove bortezomib resistance.
Conclusions: In summary, our findings indicate that artificially synthesized MIR4726EccDNA is functional in cells and that MIR4726EccDNA enhances tumor progression and partially mediates drug resistance by enhancing MIR4726-5p/NXF1/NKIRAS2 axis dependent autophagy.
Keywords: Autophagy; Bortezomib; Extrachromosomal circular DNA (EccDNA); MIR4726-5p; Multiple myeloma; NKIRAS2; NXF1.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This research was approved by the Ethics Committee of Central South University (CSU-2024-0341), and written informed consent was obtained from all patients before enrollment in the research program. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Targeting the HuR/E2F7 axis synergizes with bortezomib against multiple myeloma.Acta Pharmacol Sin. 2025 Aug;46(8):2296-2309. doi: 10.1038/s41401-025-01529-3. Epub 2025 Mar 25. Acta Pharmacol Sin. 2025. PMID: 40133626 Free PMC article.
-
Loss of GABARAP mediates resistance to immunogenic chemotherapy in multiple myeloma.Blood. 2024 Jun 20;143(25):2612-2626. doi: 10.1182/blood.2023022777. Blood. 2024. PMID: 38551812 Free PMC article.
-
RAN contributes to bortezomib resistance in multiple myeloma via regulating the Wnt/PCP pathway.Neoplasma. 2025 Jun;72(3):153-163. doi: 10.4149/neo_2025_250207N65. Neoplasma. 2025. PMID: 40635507
-
The clinical effectiveness and cost-effectiveness of bortezomib and thalidomide in combination regimens with an alkylating agent and a corticosteroid for the first-line treatment of multiple myeloma: a systematic review and economic evaluation.Health Technol Assess. 2011 Dec;15(41):1-204. doi: 10.3310/hta15410. Health Technol Assess. 2011. PMID: 22146234 Free PMC article.
-
Bortezomib for the treatment of multiple myeloma.Cochrane Database Syst Rev. 2016 Apr 20;4(4):CD010816. doi: 10.1002/14651858.CD010816.pub2. Cochrane Database Syst Rev. 2016. PMID: 27096326 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

