Temperature-dependent funnel-like DNA folding landscapes
- PMID: 40682825
- PMCID: PMC12276014
- DOI: 10.1093/nar/gkaf698
Temperature-dependent funnel-like DNA folding landscapes
Abstract
Nucleic acid hybridization in bimolecular and folding reactions is a fundamental kinetic process susceptible to water solvation, counterions, and chemical modifications with intricate enthalpy-entropy compensation effects. Such effects hinder the typically weak temperature dependencies of enthalpies and entropies quantified by the heat capacity change upon duplex formation. Using a temperature-jump optical trap, we investigate the folding thermodynamics and kinetics of DNA hairpins of varying stem sequences and loop sizes in the temperature range of 5-40○C. From a kinetic analysis and using a Clausius-Clapeyron equation in force, we derive the hybridization heat capacity changes ΔCp per GC and AT bp, finding 36 ± 3 and 29 ± 3 cal/(mol K), respectively. The almost equal values imply similar degrees of freedom arrest upon GC and AT bp formation during duplex formation. Folding kinetics on DNA hairpins of varying loop sizes show that the transition states (TS) in duplex formation have high free energies but low ΔCp values relative to the native state. Consequently, TS have low configurational entropy in agreement with the funnel-like energy landscape hypotheses. Our study underlines the validity of general principles in the hybridization and folding of nucleic acids determined by the TS's ΔCp values.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures

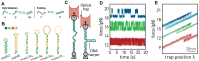
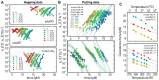

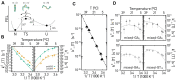

References
-
- Juraszek J, Vreede J, Bolhuis PG Transition path sampling of protein conformational changes. Chem Phys. 2012; 396:30–44. 10.1016/j.chemphys.2011.04.032. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

