The engineering of TBBPA-degrading synthetic microbiomes with integrated strategies
- PMID: 40683886
- PMCID: PMC12276246
- DOI: 10.1038/s41522-025-00777-9
The engineering of TBBPA-degrading synthetic microbiomes with integrated strategies
Abstract
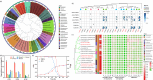
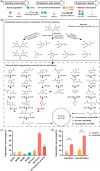
The capability to understand and construct synthetic microbiomes is crucial in biotechnological innovation and application. Tetrabromobisphenol A (TBBPA) is an emerging pollutant, and the understanding of its biodegradation is very limited. Here, a top-down approach was applied for the enrichment of TBBPA-degrading microbiomes from natural microbiomes. Ten keystone taxa correlated to TBBPA degradation and their co-occurrence interactions were identified by the dissection of the degrading microbiomes. Those keystone taxa were targeted and cultivated, and the genomic information was obtained by genome sequencing of strains and metagenomic binning. The keystone bacterial strains showed efficient degradation of TBBPA, and L-amino acids were important co-metabolic substrates to promote the degradation. Guided by this knowledge, a bottom-up approach was applied to design and construct a simplified synthetic consortium SynCon2, that consisted of four strains. The SynCon2 demonstrated efficient TBBPA degradation activity and soil bioremediation. Our study demonstrates the importance of the application of multiple tools in understanding the functions of microbiomes and provides an integrated top-down and bottom-up strategy for the construction of synthetic microbiomes with various applications.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
High-resolution mass spectrometry recognized Tetrabromobisphenol A bis (2,3-dibromo-2-methylpropyl ether) (TBBPA-DBMPE) as a contaminant in sediment from a flame retardant manufacturing factory.Water Res. 2025 Sep 1;283:123783. doi: 10.1016/j.watres.2025.123783. Epub 2025 May 4. Water Res. 2025. PMID: 40373377
-
Isolation of Monocrotophos degrading bacterial consortium from agricultural soil for in vivo analysis of pesticide degradation.Braz J Microbiol. 2024 Dec;55(4):4101-4114. doi: 10.1007/s42770-024-01497-6. Epub 2024 Sep 2. Braz J Microbiol. 2024. PMID: 39222219
-
A comparative assesment of biostimulants in microbiome-based ecorestoration of polycyclic aromatic hydrocarbon polluted soil.Braz J Microbiol. 2025 Mar;56(1):203-224. doi: 10.1007/s42770-024-01556-y. Epub 2024 Nov 27. Braz J Microbiol. 2025. PMID: 39602070
-
Keystone species in microbial communities: From discovery to soil heavy metal-remediation.J Hazard Mater. 2025 Aug 15;494:138753. doi: 10.1016/j.jhazmat.2025.138753. Epub 2025 May 27. J Hazard Mater. 2025. PMID: 40446379 Review.
-
Inhaled mannitol for cystic fibrosis.Cochrane Database Syst Rev. 2018 Feb 9;2(2):CD008649. doi: 10.1002/14651858.CD008649.pub3. Cochrane Database Syst Rev. 2018. Update in: Cochrane Database Syst Rev. 2020 May 1;5:CD008649. doi: 10.1002/14651858.CD008649.pub4. PMID: 29424930 Free PMC article. Updated.
References
-
- Philippot, L., Raaijmakers, J. M., Lemanceau, P. & van der Putten, W. H. Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol.11, 789–799 (2013). - PubMed
MeSH terms
Substances
Grants and funding
- 41991333/National Natural Science Foundation of China
- 41991333/National Natural Science Foundation of China
- 41991333/National Natural Science Foundation of China
- 41991333/National Natural Science Foundation of China
- 32270084/National Natural Science Foundation of China
- 41991333/National Natural Science Foundation of China
- 2019YFA0905500/National Key Research and Development Program of China
- 2019YFA0905500/National Key Research and Development Program of China
- 2019YFA0905500/National Key Research and Development Program of China
- 2019YFA0905500/National Key Research and Development Program of China
- 2019YFA0905500/National Key Research and Development Program of China
LinkOut - more resources
Full Text Sources

