OICR-41103 as a chemical probe for the DCAF1 WD40 domain
- PMID: 40683980
- PMCID: PMC12276300
- DOI: 10.1038/s42003-025-08491-0
OICR-41103 as a chemical probe for the DCAF1 WD40 domain
Abstract
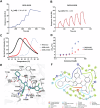
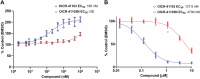
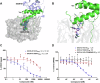
Human DCAF1 is a multidomain protein that plays a critical role in protein homeostasis. Its WDR domain functions as a substrate recruitment module for RING-type CRL4 and HECT family EDVP E3 ubiquitin ligases, enabling the ubiquitination and proteasomal degradation of specific substrates. DCAF1's activity has been implicated in cell proliferation and is documented to promote tumorigenesis. Additionally, the DCAF1 WDR domain is hijacked by lentiviral accessory proteins to induce the degradation of host antiviral factors, such as SAMHD1 and UNG2. These diverse roles make DCAF1 an attractive target for therapeutic development in oncology and antiviral strategies. It is also a promising candidate for use in targeted protein degradation. We previously reported a novel ligand, OICR-8268, that targets the DCAF1 WDR domain. In this study, we present the development of OICR-41103, a potent, selective, and cell-active small molecule chemical probe for DCAF1, derived from OICR-8268. The co-crystal structure of the DCAF1-OICR-41103 complex reveals the ligand's binding mode within the WDR central pocket, demonstrating its potential for PROTAC design and development. Notably, OICR-41103 effectively displaces the lentiviral Vpr protein from DCAF1 in both biochemical and cellular settings, highlighting its potential for the development of HIV therapeutics.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Angers, S. et al. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature443, 590–593 (2006). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

