Genotypic, functional, and phenotypic characterization in CTNNB1 neurodevelopmental syndrome
- PMID: 40684264
- PMCID: PMC12335994
- DOI: 10.1016/j.xhgg.2025.100483
Genotypic, functional, and phenotypic characterization in CTNNB1 neurodevelopmental syndrome
Abstract
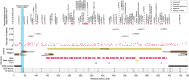
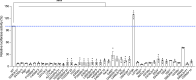
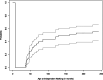
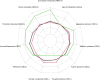
CTNNB1 neurodevelopmental syndrome is a rare disorder caused by de novo heterozygous variants in the CTNNB1 gene encoding β-catenin. This study aimed to characterize genetic variants in individuals with CTNNB1 neurodevelopmental syndrome, systematically assess the spectrum of clinical phenotypes using standardized measures, and explore potential genotype-phenotype correlations. In this cross-sectional cohort study, individuals diagnosed with CTNNB1 neurodevelopmental syndrome underwent structured interviews using standardized scales to evaluate motor skills, speech, communication, feeding abilities, visual function, neurodevelopment, and psychopathology. Genetic variants were analyzed, and, in a subset of cases, the impact of β-catenin variants on the Wnt/β-catenin signaling pathway was assessed. Across the 127 included participants (mean age, 70 months; range, 7-242 months) from 20 countries, we identified 88 different variants of the CTNNB1 gene, 87 of which were predicted to lead to loss of CTNNB1 function. Functional assays demonstrated reduced Wnt signaling activity, including 11 variants that also exhibited a dominant-negative effect. One missense variant demonstrated a gain-of-function effect. Dominant-negative variants were not clearly associated with a distinct phenotype; however, those with missense variants presented a milder phenotype, including earlier achievement of independent walking, fewer motor impairments, better conceptual and social skills, improved communication, and fewer feeding difficulties. This study describes the genetic, functional, and phenotypic characteristics in individuals with CTNNB1 neurodevelopmental syndrome. Further investigation into the genotypic and phenotypic characteristics of this syndrome and their interrelationships is essential to deepen our understanding of the disorder and inform the development of targeted therapies.
Keywords: CTNNB1 neurodevelopmental syndrome; Wnt signaling pathway; autism; developmental delay; genotype; genotype-phenotype correlations; microcephaly; phenotype; β-catenin.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no conflict of interest.
Figures






References
-
- AmeliMojarad M., AmeliMojarad M., Cui X., shariati P. Pan-cancer analysis of CTNNB1 with potential as a therapeutic target for human tumorigenesis. Inform. Med. Unlocked. 2023;42
-
- de Ligt J., Willemsen M.H., van Bon B.W.M., Kleefstra T., Yntema H.G., Kroes T., Vulto-van Silfhout A.T., Koolen D.A., de Vries P., Gilissen C., et al. Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 2012;367:1921–1929. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

