Comprehensive genotype-phenotype analysis in POLR3-related disorders
- PMID: 40684265
- PMCID: PMC12391809
- DOI: 10.1016/j.xhgg.2025.100481
Comprehensive genotype-phenotype analysis in POLR3-related disorders
Abstract
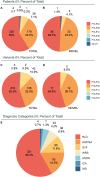
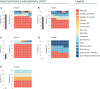
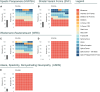
RNA polymerase III (RNA Pol III)-related disorders (POLR3-RDs) are a group of clinical entities characterized by causal variants in genes encoding RNA Pol III subunits, including POLR3A, POLR3B, POLR1C, POLR1D, POLR3D, POLR3E, POLR3F, POLR3GL, POLR3H, and POLR3K. These typically cause developmental phenotypes affecting the central nervous system; the eyes; connective tissues including bones, teeth, and endocrine axes; and the reproductive system. Similar phenotypes can be caused by variants in separate subunit genes (multigenic). In contrast, variants in the same gene can cause different phenotypes (pleiotropy), making genotype-phenotype correlation challenging. POLR3-RDs, though individually rare, have never been analyzed collectively. To bridge this gap, we developed an extensive database encompassing all published and unpublished cases of POLR3-RDs and conducted the first comprehensive genotype-phenotype correlation study across their entire spectrum. This work contributed new cases, representing 13% of all documented cases in the literature, along with 31 novel variants, accounting for 8% of all identified variants. This database was constructed by systematically reviewing the literature and integrating data from patients under the care of our international network of collaborators. The dataset includes genotype curation, bioinformatics, prior publications, and individual patient outcome information. By leveraging these comprehensive data, we were able to establish clear genotype-phenotype correlations for some pathogenic variants, which will help provide optimal clinical care and genetic counseling (including insights into disease phenotypes and progression) and offer valuable guidance for future clinical trial design and patient stratification.
Keywords: 4H leukodystrophy; POLR3-HLD; POLR3-RD; POLR3-related disorders; POLR3-related leukodystrophy; RNA Pol III; RNA polymerase III; genotype-phenotype correlations; hypodontia; hypogonadotropic hypogonadism; hypomyelination.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M.M. serves on the medical advisory board of the German Heredo Ataxia Society (DHAG) and received honoraria from Biogen, unrelated to this research. G.B. is/was a consultant for Calico (2023–present), Orchard Therapeutics (2023), Passage Bio, Inc. (2020–2022), and Ionis (2019). She is/was a site investigator/sub-I for trials and natural histories of Calico/Abbvie (2024–present), Ionis (2021–present), Shire/Takeda (2020–2021), Passage Bio (2021–2024), Bluebird Bio (2019), Regenxbio (2021–present), and Denali (2022–present). She has received an unrestricted educational grant from Takeda (2021–2022). She serves on the Scientific Advisory board of the Pelizaeus-Merzbacher Foundation, the Yaya Foundation Scientific and Clinical Advisory Council, and the Medical and Scientific Advisory Board of the United Leukodystrophy Foundation.
Figures


References
-
- Dauwerse J.G., Dixon J., Seland S., Ruivenkamp C.A.L., van Haeringen A., Hoefsloot L.H., Peters D.J.M., Boers A.C.d., Daumer-Haas C., Maiwald R., et al. Mutations in genes encoding subunits of RNA polymerases I and III cause Treacher Collins syndrome. Nat. Genet. 2011;43:20–22. - PubMed
-
- Di Bella D., Magri S., Benzoni C., Farina L., Maccagnano C., Sarto E., Moscatelli M., Baratta S., Ciano C., Piacentini S.H.M.J., et al. Hypomyelinating leukodystrophies in adults: Clinical and genetic features. Eur. J. Neurol. 2021;28:934–944. - PubMed
-
- Geschwind M., Kim M.O., Wahl C., Gahl W., Toro C. Neurology Conference: 68th American Academy of Neurology Annual Meeting, AAN 86. 2016. A novel POLR1C mutation causing autosomal recessive adult-onset leukodystrophy.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

