The antigen presentation landscape of cytokine-stressed human pancreatic islets
- PMID: 40684438
- PMCID: PMC12624573
- DOI: 10.1016/j.celrep.2025.115927
The antigen presentation landscape of cytokine-stressed human pancreatic islets
Abstract
Type 1 diabetes (T1D) arises from T cell-mediated destruction of insulin-secreting pancreatic β cells. Inflammatory triggers have been hypothesized to induce presentation of new epitopes for pathogenic T cells, but the naturally processed MHC-bound peptides presented by primary human islet β cells are largely unknown. We used mass spectrometry to identify native and post-translationally modified self-peptides presented by MHC proteins from human cadaveric islet samples treated in vitro with cytokines to identify epitopes in an inflamed pancreas. Of >4,300 islet peptides presented by 60 different MHC molecules, we identified 28 autoimmune epitopes targeted by T cells from patients with T1D, 31 additional epitopes from previously identified autoantigens, and 100 additional candidate autoantigens. The epitopes derive from inflammation, unfolded protein response, and secretory hormone processing pathways. These results identify naturally processed islet peptides targeted by autoimmune T cells in T1D and provide a resource for investigating T1D etiology and progression.
Keywords: CP: Immunology; CP: Metabolism; MHC protein; T cell epitope discovery; autoimmunity; immunopeptidome; islets of Langerhans; peptide elution; type 1 diabetes.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
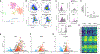



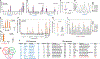
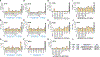
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

