Differential expression pattern of CC chemokine receptor 7 guides precision treatment of hepatocellular carcinoma
- PMID: 40685403
- PMCID: PMC12277428
- DOI: 10.1038/s41392-025-02308-6
Differential expression pattern of CC chemokine receptor 7 guides precision treatment of hepatocellular carcinoma
Abstract
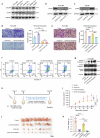
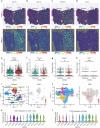
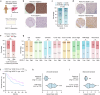
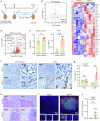
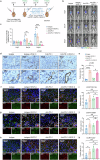
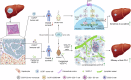
The treatment of hepatocellular carcinoma (HCC) faces challenges of low response rates to targeted drugs and immune checkpoint inhibitors, which are influenced by complicated microenvironment of HCC. In this study, the complex tumor microenvironment was identified by using tissue microarray (TMA), spatial transcriptomes and single-cell sequencing. High expression of CC chemokine receptor 7 (CCR7) in tumor cells predicted lower Overall Survival (OS). Conversely, CRISPR-Cas9-mediated knockout of CCR7 enhanced the sensitivity of HCC to sorafenib in preclinical experiments, resulting from the inhibition of epithelial-mesenchymal transition through the AKT and ERK signaling pathways. Simultaneously, we revealed CCR7 expression in stromal cells, with increased infiltration of CCR7+ immune cells into the tumor mesenchyme associated with high CCL21 expression at tumor sites. Subsequently, VEGF-C was identified as an independent predictor of higher patient OS and showed a significant positive correlation with CCR7 signaling. Interestingly, exogenous VEGF-C was found to promote the formation of tertiary lymphoid structures (TLSs) by activating lymphatic angiogenesis and the CCL21/CCR7 axis. As a result, VEGF-C treatment enhanced the efficacy of anti-PD-1 immunotherapy. This study highlights the opposing effects of tumor cell-derived versus stromal cell-derived CCR7 expression and guides the precision treatment for HCC.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

References
-
- Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin.71, 209–249 (2021). - PubMed
-
- Yang, C. et al. Evolving therapeutic landscape of advanced hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol.20, 203–222 (2023). - PubMed
-
- Pinter, M., Scheiner, B. & Pinato, D. J. Immune checkpoint inhibitors in hepatocellular carcinoma: emerging challenges in clinical practice. Lancet Gastroenterol. Hepatol.8, 760–770 (2023). - PubMed
-
- Cheng, A. L. et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol.10, 25–34 (2009). - PubMed
MeSH terms
Substances
Grants and funding
- 31471147/National Natural Science Foundation of China (National Science Foundation of China)
- 31971111/National Natural Science Foundation of China (National Science Foundation of China)
- U24A20729/National Natural Science Foundation of China (National Science Foundation of China)
- 32270886/National Natural Science Foundation of China (National Science Foundation of China)
- 82090053/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

