Blood-borne immune cells carry low biomass DNA remnants of microbes in patients with colorectal cancer or inflammatory bowel disease
- PMID: 40685618
- PMCID: PMC12283013
- DOI: 10.1080/19490976.2025.2530157
Blood-borne immune cells carry low biomass DNA remnants of microbes in patients with colorectal cancer or inflammatory bowel disease
Abstract
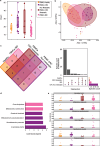
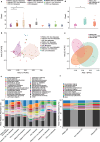
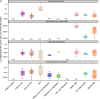
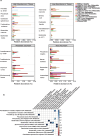
The involvement of the intestinal microbiome in the pathogenesis of inflammatory bowel disease (IBD) and colorectal cancer (CRC), is well-established. Bacteria interact with immune cells at sites of intestinal inflammation, but also in the CRC tumor microenvironment. We hypothesized that bacterial remnants translocate within peripheral blood mononuclear cells (PBMCs) into the circulation and thus explored the composition of the detectable microbiome in PBMCs of patients with CRC or IBD compared to healthy controls. The PBMC microbiome profiles partially align with the tumor-derived or intestinal tissue-derived microbiome signatures obtained from the same patients with CRC or IBD, respectively. Our metagenomics data, supported by 16S-rRNA-FISH-Flow, imaging flow cytometry and species-specific qPCR, revealed the presence of translocated bacterial genetic sequences in the patients with CRC and IBD. Thus, our data suggest that in patients with intestinal barrier leakage, there is the potential for the translocation of bacterial remnants into the circulation via PBMCs.
Keywords: Microbiome; cancer and microbiome; colorectal cancer metastasis; inflammatory bowel disease pathogenesis; intestinal epithelial barrier defect; microbiome host interaction; peripheral blood mononuclear cells.
Conflict of interest statement
MS has shares and is co-founder of Recolony AG, Zurich, CH and has shares in PharmaBiome AG, Zurich, CH. MS served as Advisor for Abbvie, Gilead, Fresenius, Topadur, Takeda, Roche, Astra Zeneca and Celltrion. MS received speaker’s honoraria from Janssen, Falk Pharma, Vifor Pharma, Pileje and Bromatech. MS received research grants from Abbvie, Takeda, Gilead, Gnubiotics, Roche, Axalbion, Pharmabiome, Topadur, Basilea, MBiomics, Storm Therapeutics, LimmatTech, Zealand Pharma, NodThera, Calypso Biotech, Menarini. Pileje, Herbodee, Vifor. GR has shares and is cofounder and head of the scientific advisory board of PharmaBiome. GR has consulted to Abbvie, Arena, Augurix, BMS, Boehringer, Calypso, Celgene, FALK, Ferring, Fisher, Genentech, Gilead, Janssen, Lilly, MSD, Novartis, Pfizer, Phadia, Roche, UCB, Takeda, Tillots, Vifor, Vital Solutions and Zeller. GR received speaker’s honoraria from Abbvie, Astra Zeneca, BMS, Celgene, FALK, Janssen, MSD, Pfizer, Phadia, Takeda, Tillots, UCB, Vifor and Zeller. GR received educational grants and research grants from Abbvie, Ardeypharm, Augurix, Calypso, FALK, Flamentera, MSD, Novartis, Pfizer, Roche, Takeda, Tillots, UCB and Zeller. MT served as advisor for Topadur and Takeda. MT received speaker’s honoraria from Janssen, Takeda and Intuitive Surgical. RF has served as an Advisor or speaker for Roche, Pierre Fabre Pharma, Servier, Bristol Myers Squibb, Merck Sharp&Dome, Astra Zeneca. LB has served as advisor for Abbvie, Amgen, BMS, Falk, Janssen, Pfizer, Lilly, Takeda, Sanofi, Esocap, Aquilion and received speaker fees from Takeda, Sanofi, Abbvie, Janssen, Lilly, Falk, BMS, Pfizer. The additional authors declare that they have no competing interests relevant to this work.
Figures



References
-
- Zhang Y, Zhang L, Zheng S, Li M, Xu C, Jia D, Qi Y, Hou T, Wang L, Wang B, et al. Fusobacterium nucleatum promotes colorectal cancer cells adhesion to endothelial cells and facilitates extravasation and metastasis by inducing ALPK1/NF-kappaB/ICAM1 axis. Gut Microbes. 2022;14(1):2038852. doi: 10.1080/19490976.2022.2038852. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
