Lung Single-Cell Transcriptomics Reveal Diverging Pathobiology and Opportunities for Precision Targeting in Scleroderma-Associated Versus Idiopathic Pulmonary Arterial Hypertension
- PMID: 40686216
- PMCID: PMC12313177
- DOI: 10.1161/CIRCGEN.124.004936
Lung Single-Cell Transcriptomics Reveal Diverging Pathobiology and Opportunities for Precision Targeting in Scleroderma-Associated Versus Idiopathic Pulmonary Arterial Hypertension
Abstract
Background: Pulmonary arterial hypertension (PAH) involves progressive cellular and molecular change within the pulmonary vasculature, leading to increased vascular resistance. Current therapies targeting nitric oxide, endothelin, and prostacyclin pathways yield variable treatment responses. Patients with systemic sclerosis-associated PAH (SSc-PAH) often experience worse outcomes than those with idiopathic PAH (IPAH). We hypothesized that distinct and overlapping gene expression patterns in SSc-PAH versus IPAH lung tissues could inform the investigation of precision-targeted therapies.
Methods: Lung tissue samples from 4 SSc-PAH, 4 IPAH, and 4 failed donor specimens were obtained from the Pulmonary Hypertension Breakthrough Initiative lung tissue bank. Single-cell RNA sequencing was performed using the 10X Genomics Chromium Flex platform. Data normalization, clustering, and differential expression analysis were conducted using Seurat. Additional analyses included gene set enrichment analysis, transcription factor activity analysis, and ligand-receptor signaling. Pharmacotranscriptomic screening was performed using the Connectivity Map.
Results: SSc-PAH samples showed a higher proportion of fibroblasts compared with failed donors and a higher proportion of dendritic cells/macrophages compared with IPAH. Gene set enrichment analysis revealed enriched pathways related to epithelial-to-mesenchymal transition, apoptosis, and vascular remodeling in SSc-PAH samples. There was pronounced differential gene expression across diverse pulmonary vascular cell types and in various epithelial cell types in both IPAH and SSc-PAH, with epithelial-to-endothelial cell signaling observed. Macrophage-to-endothelial cell signaling was particularly pronounced in SSc-PAH. Pharmacotranscriptomic screening identified TIE2, GSK-3, and PKC inhibitors, among other compounds, as potential drug candidates for reversing SSc-PAH gene expression signatures.
Conclusions: Overlapping and distinct gene expression patterns exist in SSc-PAH versus IPAH, with significant molecular differences suggesting unique pathogenic mechanisms in SSc-PAH. These findings highlight the potential for precision-targeted therapies to improve outcomes in patient with SSc-PAH. Future studies should validate these targets and explore their therapeutic efficacy.
Keywords: computational biology; lung; precision medicine; pulmonary arterial hypertension; scleroderma, systemic; transcriptome.
Conflict of interest statement
Dr Cherry is the owner of C M Cherry Consulting. The other authors report no conflicts.
Figures



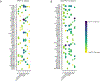
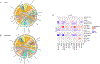

Update of
-
Single-cell transcriptomics reveal diverging pathobiology and opportunities for precision targeting in scleroderma-associated versus idiopathic pulmonary arterial hypertension.bioRxiv [Preprint]. 2024 Oct 25:2024.10.25.620225. doi: 10.1101/2024.10.25.620225. bioRxiv. 2024. Update in: Circ Genom Precis Med. 2025 Aug;18(4):e004936. doi: 10.1161/CIRCGEN.124.004936. PMID: 39484590 Free PMC article. Updated. Preprint.
References
-
- Hassoun PM. Pulmonary Arterial Hypertension. N Engl J Med. 2021;385:2361–2376. - PubMed
-
- Humbert M, Kovacs G, Hoeper MM, Badagliacca R, Berger RMF, Brida M, Carlsen J, Coats AJS, Escribano-Subias P, Ferrari P, Ferreira DS, Ghofrani HA, Giannakoulas G, Kiely DG, Mayer E, Meszaros G, Nagavci B, Olsson KM, Pepke-Zaba J, Quint JK, Radegran G, Simonneau G, Sitbon O, Tonia T, Toshner M, Vachiery JL, Vonk Noordegraaf A, Delcroix M, Rosenkranz S and Group EESD. 2022 ESC/ERS Guidelines for the diagnosis and treatment of pulmonary hypertension. Eur Respir J. 2022. - PubMed
-
- Humbert M, Sitbon O, Guignabert C, Savale L, Boucly A, Gallant-Dewavrin M, McLaughlin V, Hoeper MM and Weatherald J. Treatment of pulmonary arterial hypertension: recent progress and a look to the future. Lancet Respir Med. 2023;11:804–819. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

