Inference of antimicrobial resistance (AMR) from a whole genome database outperforming AMR gene detection
- PMID: 40686603
- PMCID: PMC12275941
- DOI: 10.1016/j.isci.2025.112962
Inference of antimicrobial resistance (AMR) from a whole genome database outperforming AMR gene detection
Abstract
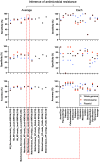
This study focuses on the rapid detection of antimicrobial resistance (AMR) in Klebsiella pneumoniae. The "Align-Search-Infer" pipeline aligned query sequences from 24 urine samples against a curated genome database of 40 Klebsiella isolates, searched for the best matches, and inferred their antimicrobial susceptibility. Carbapenem resistance inference achieved 77.3% accuracy (95%CI: 59.8-94.8%) within 10 min using whole-genome matching, and 85.7% accuracy (95%CI: 70.7-100.0%) within 1 h using plasmid matching - both surpassing the 54.2% accuracy (95%CI: 34.2-74.1%) of AMR gene detection at 6 h. The proposed method requires less bacterial DNA and is suitable for low-load clinical samples. Our small local database performed comparably to large public databases. This study supports the integration of pathogen-specific genome databases into clinical workflows to enable rapid and accurate antimicrobial susceptibility prediction. Further research is needed to validate and refine the method using larger genomic-phenotypic datasets across diverse pathogens and sample types.
Keywords: Biological sciences; Microbial genomics; Microbiology; Natural sciences.
© 2025 The Authors.
Conflict of interest statement
The authors declared no conflict of interest.
Figures






References
LinkOut - more resources
Full Text Sources

