Analysis of metagenomic data
- PMID: 40688383
- PMCID: PMC12276902
- DOI: 10.1038/s43586-024-00376-6
Analysis of metagenomic data
Abstract
Metagenomics has revolutionized our understanding of microbial communities, offering unprecedented insights into their genetic and functional diversity across Earth's diverse ecosystems. Beyond their roles as environmental constituents, microbiomes act as symbionts, profoundly influencing the health and function of their host organisms. Given the inherent complexity of these communities and the diverse environments where they reside, the components of a metagenomics study must be carefully tailored to yield accurate results that are representative of the populations of interest. This Primer article examines the methodological advancements and current practices that have shaped the field, from initial stages of sample collection and DNA extraction to the advanced bioinformatics tools employed for data analysis, with a particular focus on the profound impact of next-generation sequencing (NGS) on the scale and accuracy of metagenomics studies. We critically assess the challenges and limitations inherent in metagenomics experimentation, available technologies and computational analysis methods. Beyond technical methodologies, we explore the application of metagenomics across various domains, including human health, agriculture and environmental monitoring. Looking ahead, we advocate for the development of more robust computational frameworks and enhanced interdisciplinary collaborations. This Primer serves as a comprehensive guide for advancing the precision and applicability of metagenomic studies, positioning them to address the complexities of microbial ecology and their broader implications for human health and environmental sustainability.
Conflict of interest statement
Competing interests The authors declare no competing interests.
Figures

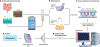


References
-
- Handelsman J, Rondon MR, Brady SF, Clardy J & Goodman RM Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol 5, R245–249 (1998). - PubMed
-
- Venter JC et al. Environmental Genome Shotgun Sequencing of the Sargasso Sea. Science 304, 66–74 (2004). - PubMed
-
- Sunagawa S et al. Tara Oceans: towards global ocean ecosystems biology. Nat Rev Microbiol 18, 428–445 (2020). - PubMed
Grants and funding
- U01 DA053941/DA/NIDA NIH HHS/United States
- R01 AI125416/AI/NIAID NIH HHS/United States
- U19 AI174998/AI/NIAID NIH HHS/United States
- U54 AG089334/AG/NIA NIH HHS/United States
- R35 GM146980/GM/NIGMS NIH HHS/United States
- R01 GM146462/GM/NIGMS NIH HHS/United States
- R01 AI173172/AI/NIAID NIH HHS/United States
- R01 AI100947/AI/NIAID NIH HHS/United States
- R21 EB031466/EB/NIBIB NIH HHS/United States
- R01 AI151059/AI/NIAID NIH HHS/United States
- R21 AI129851/AI/NIAID NIH HHS/United States
- R35 GM138369/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
