Molecular characterization of the oriental cat flea, Ctenocephalides orientis, parasitizing goats based on nuclear and mitochondrial protein-coding genes
- PMID: 40689040
- PMCID: PMC12272463
- DOI: 10.1016/j.crpvbd.2025.100286
Molecular characterization of the oriental cat flea, Ctenocephalides orientis, parasitizing goats based on nuclear and mitochondrial protein-coding genes
Abstract
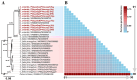
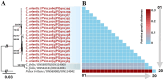
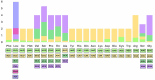
Fleas are wingless ectoparasites that feed on the blood of warm-blooded animals and play a significant role as vectors of several medically and veterinary-relevant diseases. The oriental cat flea, Ctenocephalides orientis, is endemic to Asia and infests dogs more frequently than cats. However, its presence in small ruminants remains largely unexplored. Between January 2017 and October 2023, flea surveys were conducted on goat farms across seven different provinces in Thailand. Initially, flea specimens were identified using morphological keys and, subsequently, confirmed through molecular analysis of the mitochondrial genes cytochrome c oxidase subunit 1 (cox1, 450 bp) and cytochrome c oxidase subunit 2 (cox2, 678 bp), the nuclear ribosomal internal transcribed spacer 1 (ITS1, 828 bp) and the elongation factor-1 alpha (EF-1α, 904 bp) gene. In addition to characterizing these markers, the mitochondrial genome, including all protein-coding genes (PCGs), was amplified, analyzed, and subjected to comparative analyses. Among 500 goats examined, 33 (6.6%) were infested with fleas, which belonged to only one species, C. orientis. Pairwise genetic distance analysis and phylogenetic reconstruction strongly supported the placement of C. orientis within a distinct clade, consistent with the reference sequences. Of the four genetic markers analyzed, EF-1α exhibited the highest diversity. The partial mitochondrial genome of C. orientalis (14,315 bp) encoding 34 genes, including 13 PCGs, 19 transfer RNA genes, and two ribosomal RNA genes, was sequenced. Phylogenetic and genetic distance analyses based on multiple molecular markers and the mitochondrial genome revealed a close evolutionary relationship between C. orientis and C. canis. These findings confirmed that C. orientis is not only restricted to companion animals but also infests goats, suggesting its potential role in disease transmission to other animals. Furthermore, the study findings provide a dataset of both nuclear and mitochondrial molecular markers, which would facilitate future research on the taxonomy, phylogeny, and evolutionary relationships of fleas.
Keywords: Ctenocephlides orientis; EF-1α; Flea; Genetic diversity; Mitochondrial genome; cox1; cox2.
© 2025 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Ashwini M., Puttalakshmamma G., Mamatha G., Chandranaik B., Thimmareddy P., Placid E., et al. Studies on morphology and molecular characterization of oriental cat flea infesting small ruminants by barcoding. J. Entomol. Zool. Stud. 2017;5:301–305.
-
- Aung A., Narapakdeesakul D., Arnuphapprasert A., Nugraheni Y.R., Wattanachant C., Kaewlamun W., et al. Multi-locus sequence analysis of Anaplasma bovis in goats and ticks from Thailand, with the initial identification of an uncultured Anaplasma species closely related to Anaplasma phagocytophilum-like 1. Comp. Immunol. Microbiol. Infect. Dis. 2024;109 - PubMed
-
- Azrizal-Wahid N., Sofian-Azirun M., Low V.L. New insights into the haplotype diversity of the cosmopolitan cat flea Ctenocephalides felis (Siphonaptera: Pulicidae) Vet. Parasitol. 2020;281 - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

