A UFD1 variant encoding a microprotein modulates UFD1f and IPMK ubiquitination to play pivotal roles in anti-stress responses
- PMID: 40691175
- PMCID: PMC12280206
- DOI: 10.1038/s41467-025-62073-6
A UFD1 variant encoding a microprotein modulates UFD1f and IPMK ubiquitination to play pivotal roles in anti-stress responses
Abstract
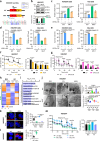
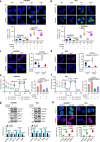
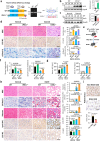
Eukaryotic cells make multiple efforts to cope with internal and external stresses; such mechanisms include metabolic responses and the generation of stress-responsive mRNA isoforms (SR-mRNAisos), such as the classical XBP1s. Here, we identified a mammalian conserved SR-mRNAiso, UFD1s, which encodes a microprotein with anti-stress functions. UFD1s decreased the K63-linked ubiquitination levels of UFD1 full-length protein (UFD1f) via competitive binding to the E3 ubiquitin ligase MARCH7, and therefore regulated the dynamics of protein ubiquitination. Inositol polyphosphate multikinase (IPMK) was identified as the most significantly UFD1s-regulated target in terms of changes in K48- and K11-ubiquitination. UFD1s promoted autophagy and fatty acid oxidation, and IPMK was consistently destabilized. Ufd1s-deficient male mice exhibited metabolic disorders and accelerated NASH progression. Plasmid or circRNA expressing UFD1s alleviated NASH in mice, indicating that UFD1s has therapeutic value. Our findings revealed a mammalian conserved microprotein that plays crucial roles in anti-stress regulation through the modulation of ubiquitination and metabolism.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare the following patent application related to this work. Patent applicants: Institute of Health and Medicine, Hefei Comprehensive National Science Center; University of Science and Technology of China. Inventors: G.S., X.Li., X.W. (co-authors of this article); Application number: CN 2024114540916; Status: Pending; The patent claims cover the application of small protein UFD1s for the prevention, treatment, and diagnosis of diseases, including NASH. The remaining authors declare no competing interests.
Figures





References
-
- Galluzzi, L., Yamazaki, T. & Kroemer, G. Linking cellular stress responses to systemic homeostasis. Nat. Rev. Mol. Cell Biol.19, 731–745 (2018). - PubMed
-
- Kevei, É. & Hoppe, T. Ubiquitin sets the timer: impacts on aging and longevity. Nat. Struct. Mol. Biol.21, 290–292 (2014). - PubMed
-
- Hetz, C., Chevet, E. & Harding, H. P. Targeting the unfolded protein response in disease. Nat. Rev. Drug Discov.12, 703–719 (2013). - PubMed
MeSH terms
Substances
Grants and funding
- 32470596/National Natural Science Foundation of China (National Science Foundation of China)
- 32000438/National Natural Science Foundation of China (National Science Foundation of China)
- U23A20164/National Natural Science Foundation of China (National Science Foundation of China)
- 32270590/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

