E2F activity determines mitosis versus whole-genome duplication in G2-arrested cells
- PMID: 40691182
- PMCID: PMC12279939
- DOI: 10.1038/s41467-025-62061-w
E2F activity determines mitosis versus whole-genome duplication in G2-arrested cells
Abstract
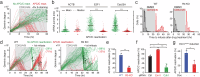
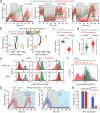
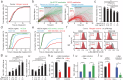
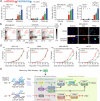
While mitogenic signaling is known to regulate cell-cycle entry during the G1 phase, its function in the G2 phase remains elusive. Here we show that mitogenic signaling controls whether G2-arrested cells proceed through mitosis or undergo whole-genome duplication. Although mitogenic signaling is not required for the G2/M transition under normal conditions, it modulates E2F transcriptional activity via c-Myc. When G2 arrest occurs due to CDK4/6 and CDK2 suppression, E2F activity levels determine the status of APC/C inactivation and the CDK2-Rb feedback loop. Upon release from G2 arrest, cells maintaining APC/C inactivation promptly induce CDK2 activation and FoxM1 phosphorylation, driving mitotic entry. Conversely, APC/C reactivation degrades cyclin A and abolishes the CDK2-Rb loop, necessitating CDK4/6 activation for cell-cycle re-entry. This regulatory mechanism mirrors the G1-phase process, resulting in whole-genome duplication. In cancer cells, this process promotes genome instability and oncogene amplification, contributing to aggressive behavior. These findings reveal a previously unrecognized mitogen-dependent checkpoint that governs cell fate in the G2 phase.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

