Aberrant Notch-signaling promotes tumor angiogenesis in esophageal squamous-cell carcinoma
- PMID: 40691441
- PMCID: PMC12280033
- DOI: 10.1038/s41392-025-02309-5
Aberrant Notch-signaling promotes tumor angiogenesis in esophageal squamous-cell carcinoma
Erratum in
-
Correction: Aberrant Notch-signaling promotes tumor angiogenesis in esophageal squamous-cell carcinoma.Signal Transduct Target Ther. 2025 Aug 31;10(1):288. doi: 10.1038/s41392-025-02403-8. Signal Transduct Target Ther. 2025. PMID: 40887484 Free PMC article. No abstract available.
Abstract
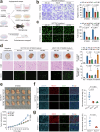
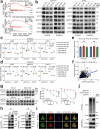
Esophageal squamous-cell carcinoma (ESCC) is one of the most common gastrointestinal cancers in China, characterized by high malignancy and poor prognosis. Nowadays, the therapeutic options for this cancer are very limited. Notch-signaling is often overactivated in ESCC, but its role remains to be fully elucidated. Here, we demonstrate that aberrant Notch-signaling plays an important role in tumor angiogenesis. In clinical ESCC samples, Notch-signaling activation scores were significantly correlated with tumor microvascular density, advanced TNM stages, and short patient survival time. Silencing Notch-signaling substantially suppressed the ability of ESCC cells to promote angiogenesis in vitro and in vivo. By integrating analysis of CUT&Tag and RNA sequencing data, we identified ubiquitin-specific protease 5 (USP5) as a Notch-signaling downstream effector that is transcriptionally upregulated by the NOTCH1 intracellular domain (NICD1)-RBPJ complex and mediates tumor angiogenesis. USP5 stabilized STAT3 via its deubiquitination function, thereby enhancing the production of pro-angiogenic factors by cancer cells, including VEGF, ANGPT2, and CXCL1. We showed that chemotherapy combined with the USP5 inhibitor can additionally repress tumor growth and angiogenesis in mice. These findings explain why ESCC cells have much fewer NOTCH1 mutations than normal and precancerous epithelium, reveal a novel mechanism for Notch-signaling to drive tumor angiogenesis via the NOTCH1-USP5-STAT3 axis, and open a potential new avenue for anti-tumor angiogenesis therapy.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

