The hit-and-run of cell wall synthesis: LpoB transiently binds and activates PBP1b through a conserved allosteric switch
- PMID: 40691462
- PMCID: PMC12279963
- DOI: 10.1038/s41467-025-62051-y
The hit-and-run of cell wall synthesis: LpoB transiently binds and activates PBP1b through a conserved allosteric switch
Abstract
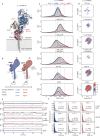
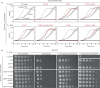
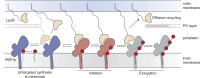
The peptidoglycan (PG) cell wall is the primary protective layer of bacteria, making the process of PG synthesis a key antibiotic target. Class A penicillin-binding proteins (aPBPs) are a family of conserved and ubiquitous PG synthases that fortify and repair the PG matrix. In gram-negative bacteria, these enzymes are regulated by outer-membrane tethered lipoproteins. However, the molecular mechanism by which lipoproteins coordinate the spatial recruitment and enzymatic activation of aPBPs remains unclear. Here we use single-molecule FRET and single-particle tracking in E. coli to show that a prototypical lipoprotein activator LpoB triggers site-specific PG synthesis by PBP1b through conformational rearrangements. Once synthesis is initiated, LpoB affinity for PBP1b dramatically decreases and it dissociates from the synthesizing enzyme. Our results suggest that transient allosteric coupling between PBP1b and LpoB directs PG synthesis to areas of low peptidoglycan density, while simultaneously facilitating efficient lipoprotein redistribution to other sites in need of fortification.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: A.C.K. is a cofounder and consultant for biotechnology companies Tectonic Therapeutic and Seismic Therapeutic, and for the Institute for Protein Innovation, a non-profit research institute. The remaining authors declare no competing interests.
Figures



Update of
-
The hit-and-run of cell wall synthesis: LpoB transiently binds and activates PBP1b through a conserved allosteric switch.bioRxiv [Preprint]. 2024 Dec 14:2024.12.13.628440. doi: 10.1101/2024.12.13.628440. bioRxiv. 2024. Update in: Nat Commun. 2025 Jul 21;16(1):6723. doi: 10.1038/s41467-025-62051-y. PMID: 39713382 Free PMC article. Updated. Preprint.
Similar articles
-
The hit-and-run of cell wall synthesis: LpoB transiently binds and activates PBP1b through a conserved allosteric switch.bioRxiv [Preprint]. 2024 Dec 14:2024.12.13.628440. doi: 10.1101/2024.12.13.628440. bioRxiv. 2024. Update in: Nat Commun. 2025 Jul 21;16(1):6723. doi: 10.1038/s41467-025-62051-y. PMID: 39713382 Free PMC article. Updated. Preprint.
-
Outer-membrane lipoprotein LpoB spans the periplasm to stimulate the peptidoglycan synthase PBP1B.Proc Natl Acad Sci U S A. 2014 Jun 3;111(22):8197-202. doi: 10.1073/pnas.1400376111. Epub 2014 May 12. Proc Natl Acad Sci U S A. 2014. PMID: 24821816 Free PMC article.
-
Induced conformational changes activate the peptidoglycan synthase PBP1B.Mol Microbiol. 2018 Nov;110(3):335-356. doi: 10.1111/mmi.14082. Epub 2018 Oct 25. Mol Microbiol. 2018. PMID: 30044025 Free PMC article.
-
Decoding the Penicillin-Binding Proteins with Activity-Based Probes.Acc Chem Res. 2025 Jun 3;58(11):1754-1763. doi: 10.1021/acs.accounts.5c00113. Epub 2025 May 21. Acc Chem Res. 2025. PMID: 40396497 Review.
-
Activities and regulation of peptidoglycan synthases.Philos Trans R Soc Lond B Biol Sci. 2015 Oct 5;370(1679):20150031. doi: 10.1098/rstb.2015.0031. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 26370943 Free PMC article. Review.
References
-
- Silver, L. L. Viable screening targets related to the bacterial cell wall. Ann. N. Y Acad. Sci.1277, 29–53 (2013). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

