A deep ensemble framework for human essential gene prediction by integrating multi-omics data
- PMID: 40691502
- PMCID: PMC12280155
- DOI: 10.1038/s41598-025-99164-9
A deep ensemble framework for human essential gene prediction by integrating multi-omics data
Abstract
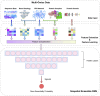
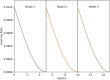
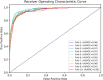
Essential genes are necessary for the survival or reproduction of a living organism. The prediction and analysis of gene essentiality can advance our understanding of basic life and human diseases, and further boost the development of new drugs. We propose a snapshot ensemble deep neural network method, DeEPsnap, to predict human essential genes. DeEPsnap integrates the features derived from DNA and protein sequence data with the features extracted or learned from four types of functional data: gene ontology, protein complex, protein domain, and protein-protein interaction networks. More than 200 features from these biological data are extracted/learned which are integrated together to train a series of cost-sensitive deep neural networks. The proposed snapshot mechanism enables us to train multiple models without increasing extra training effort and cost. The experimental results of 10-fold cross-validation show that DeEPsnap can accurately predict human gene essentiality with an average AUROC of 96.16%, AUPRC of 93.83%, and accuracy of 92.36%. The comparative experiments show that DeEPsnap outperforms several popular traditional machine learning models and deep learning models, while all those models show promising performance using the features we created for DeEPsnap. We demonstrated that the proposed method, DeEPsnap, is effective for predicting human essential genes.
Keywords: Deep learning; Essential gene prediction; Multi-omics data integration; Snapshot ensemble.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors have declared that no competing interests exist.
Figures
Similar articles
-
Development and Validation of a Convolutional Neural Network Model to Predict a Pathologic Fracture in the Proximal Femur Using Abdomen and Pelvis CT Images of Patients With Advanced Cancer.Clin Orthop Relat Res. 2023 Nov 1;481(11):2247-2256. doi: 10.1097/CORR.0000000000002771. Epub 2023 Aug 23. Clin Orthop Relat Res. 2023. PMID: 37615504 Free PMC article.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Artificial intelligence for diagnosing exudative age-related macular degeneration.Cochrane Database Syst Rev. 2024 Oct 17;10(10):CD015522. doi: 10.1002/14651858.CD015522.pub2. Cochrane Database Syst Rev. 2024. PMID: 39417312
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
-
- Hart, T. et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell163, 1515–1526. 10.1016/j.cell.2015.11.015 (2015). - PubMed
-
- Liao, B. & Zhang, J. Mouse duplicate genes are as essential as singletons. Trends Genet.23, 378–381. 10.1016/j.tig.2007.05.006 (2007). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

