Genome-wide identification of key genes related to chloride ion (Cl-) channels and transporters in response to salt stress in birch
- PMID: 40691768
- PMCID: PMC12278563
- DOI: 10.1186/s12864-025-11795-4
Genome-wide identification of key genes related to chloride ion (Cl-) channels and transporters in response to salt stress in birch
Abstract
Background: Soil salinization is a common matter of concern all over the world, which severely affects plant production, soil health, and ecosystem stability. Birch is a significant afforestation tree species with great ecological, economic, and evolutionary value. It is an excellent model system for studying the acclimation and adaptation of woody plants to extreme environmental conditions due to the current advancements in genomics, genetic variability, and extensive studies with a focus on various biotic and abiotic stresses. To date, the genetic regulation of birch trees in defending against environmental stimuli, particularly salt stress, has made great progress. However, the information on how genes related to chloride ion (Cl-) channels and transporters respond to salt stress remains poorly understood in birch.
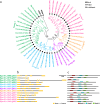
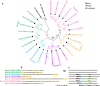
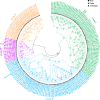
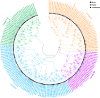
Results: Herein, we performed the genome-wide identification, evolutionary relationship, sequence analysis, and expression patterns of seven family genes encoding Cl- channels or transporters in birch. We identified one, one, four, seven, 13, 56, and 67 genes belonging to the cation chloride co-transporter (CCC) family, bestrophin family, slow type anion channel (SLAC) family, chloride channel (CLC) family, aluminum-activated malate transporter (ALMT) family, nitrate transporter 1/peptide transporter family (NPF) family, and multidrug and toxic compound extrusion (MATE ) family, respectively. Except for motif prediction, a high similarity of the conserved domain composition, gene structures, and physicochemical properties was observed within each group of a certain gene family. Gene Ontology (GO) classification analysis showed that the genes encoding ALMTs, CCCs, CLCs, MATEs, NPFs, and SLACs were mainly involved in ion transport. Transcriptome profiles showed that there were 52 genes related to Cl- transporters and channels differentially expressed in any tissue of the two birch species subjected to salt treatment. Of these, three genes, designated as BpCLCa/b, BpDTX45, and BpDTX18/19, were simultaneously expressed at altered levels in all tissues of the two birch species, which was consistent with the results of quantitative real-time polymerase chain reaction (RT-qPCR) assay. In addition, the molecular docking simulations showed that sodium ion (Na+) and Cl-, respectively, had a strong binding affinity towards BpCLCa/b, BpDTX45, and BpDTX18/19.
Conclusion: A total of 149 genes related to Cl- channels and transporters were identified from the birch genome. We proposed BpCLCa/b, BpDTX45, and BpDTX18/19 as ion channels or transporters in controlling ionic homeostasis during salt stress response. The findings of our study can provide valuable resources for genetic improvement of salt-tolerant birch varieties for high-quality plantation forests in harsh environments.
Keywords: Birch; Chlorine; Ion channel; Salt stress; Sodium; Transporter.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study was carried out in accordance with the People’s Republic of China and international authorities relevant guidelines and legislation, including the official website of the Committee on Publication Ethics ( http://www.publicationethics.org/ ) and the European Association of Science Editors (EASE) and other institutions’ publishing ethics standards. All plants materials occurred in the present study are used for scientific research, which are allowed to be used and provided free of charge. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Genome-wide characterization of GRAS gene family and their expression profiles under diverse biotic and abiotic stresses in Amorphophallus konjac.BMC Genomics. 2025 Jul 8;26(1):643. doi: 10.1186/s12864-025-11777-6. BMC Genomics. 2025. PMID: 40629278 Free PMC article.
-
The effect of sample site and collection procedure on identification of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Dec 16;12(12):CD014780. doi: 10.1002/14651858.CD014780. Cochrane Database Syst Rev. 2024. PMID: 39679851 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Drugs for preventing postoperative nausea and vomiting in adults after general anaesthesia: a network meta-analysis.Cochrane Database Syst Rev. 2020 Oct 19;10(10):CD012859. doi: 10.1002/14651858.CD012859.pub2. Cochrane Database Syst Rev. 2020. PMID: 33075160 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

