Respiratory microbiome alterations, coinfections and virus intra-host evolution in a persistently active SARS-CoV-2 infection
- PMID: 40691821
- PMCID: PMC12281699
- DOI: 10.1186/s12879-025-11355-x
Respiratory microbiome alterations, coinfections and virus intra-host evolution in a persistently active SARS-CoV-2 infection
Abstract
Background: Respiratory microbiome alterations, coinfections, and virus intrahost evolution are of great interest in persistently viable SARS-CoV-2 infections in the context of antiviral treatment and immune response. However, samples before, during, and after infection are seldom available to researchers. Therefore, there has been a significant lack of opportunities to comprehensively study microbiota homeostasis, coinfections, and virus intra-host evolution on the consensus and minor variants scale in response to antiviral treatments and patient immune response.
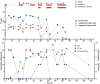
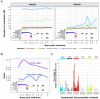
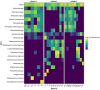
Case presentation: A 63-year-old female patient with diffuse large B-cell lymphoma received multiple treatments for SARS-CoV-2 that remained active 169 days. Together, 32 respiratory and 19 serum samples were collected before, during, and after (- 398 to 233 days) COVID-19. Subsets were selected for virus viability testing by culture (20) and subgenomic (sg) RNA (20) measurement, intra-host evolution assessment (18), microbiome composition analysis (28), and coinfection identification (11). IgA/IgG and neutralizing anti-SARS-CoV-2 antibodies were measured 19 times throughout the infection. SARSCoV-2 lineage XBB.1.16.11 persisted and remained viable until 116 days post infection (PI) regardless of treatments. No sgRNA marker tested was suitable for virus viability prediction. IgG/IgA antibodies first appeared after 38 days, but the virus persisted regardless of multiple plasma treatments before neutralizing antibodies appeared (100 days PI) and finally cleared the virus 116 days PI. Consensus-level mutations fluctuated around 102.7 ± 4.0, and minor variants increased from six to 61 with a mutation rate of 4.9 × 10-3 per site per year, with the highest average number of mutations per gene length in S and E (0.013) with surges after every antiviral treatment. The transversion/transition ratio increased from 0.50 (day 0) to 0.57 (day 24) with a steady decrease to 0.48 (day 147). Mutational signature analysis showed dominance of C > T substitutions consistent with APOBEC antiviral enzyme activity. Upper respiratory microbiota showed three distinct profiles with varying α-/β-diversity and an association of Staphylococcus spp. with COVID-19.
Conclusions: These findings further elucidate the dynamics of intra-host viral evolution and complexities of virus clearance in individuals with hematological malignancies and highlight the impact of antiviral treatments on the potential of virus variants emergence in longitudinally infectious patients due to delayed immune response.
Keywords: B-cell lymphoma; Hematological malignancies; Microbiome; Prolonged SARS-CoV-2 infection; Virus evolution; Virus shedding.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study was performed in accordance with the ethical guidelines for human research, the World Medical Association’s Declaration of Helsinki, the Oviedo Convention on Human Rights and Biomedicine, and the Slovenian Code of Medical Deontology. The study was approved by the Institutional Ethics Committee (ERIDNPVO-0052/2024). Consent for publication: A written informed consent was obtained by the patient for the publication of this case report. Competing interests: The authors declare no competing interests.
Figures
Similar articles
-
Virological Aspects of COVID-19 in Patients with Hematological Malignancies: Duration of Viral Shedding and Genetic Analysis.Viruses. 2024 Dec 31;17(1):46. doi: 10.3390/v17010046. Viruses. 2024. PMID: 39861838 Free PMC article.
-
Antibody tests for identification of current and past infection with SARS-CoV-2.Cochrane Database Syst Rev. 2022 Nov 17;11(11):CD013652. doi: 10.1002/14651858.CD013652.pub2. Cochrane Database Syst Rev. 2022. PMID: 36394900 Free PMC article.
-
SARS-CoV-2-neutralising monoclonal antibodies to prevent COVID-19.Cochrane Database Syst Rev. 2022 Jun 17;6(6):CD014945. doi: 10.1002/14651858.CD014945.pub2. Cochrane Database Syst Rev. 2022. PMID: 35713300 Free PMC article.
-
Physical interventions to interrupt or reduce the spread of respiratory viruses.Cochrane Database Syst Rev. 2023 Jan 30;1(1):CD006207. doi: 10.1002/14651858.CD006207.pub6. Cochrane Database Syst Rev. 2023. PMID: 36715243 Free PMC article.
-
COVID-19 Vaccines.2025 Jul 15. Drugs and Lactation Database (LactMed®) [Internet]. Bethesda (MD): National Institute of Child Health and Human Development; 2006–. 2025 Jul 15. Drugs and Lactation Database (LactMed®) [Internet]. Bethesda (MD): National Institute of Child Health and Human Development; 2006–. PMID: 33355732 Free Books & Documents. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- P3-0289/The Slovenian Research and Innovation Agency
- P3-0083, J3-50101/The Slovenian Research and Innovation Agency
- P3-0083, J3-50101/The Slovenian Research and Innovation Agency
- P3-0083, J3-50101/The Slovenian Research and Innovation Agency
- P3-0083, J3-50101/The Slovenian Research and Innovation Agency
- P3-0083, J3-50101/The Slovenian Research and Innovation Agency
- P3-0083, J3-50101/The Slovenian Research and Innovation Agency
- MRIC-UL-IC-BSL3+, IP-022/Univerza v Ljubljani
- MRIC-UL-IC-BSL3+, IP-022/Univerza v Ljubljani
- MRIC-UL-IC-BSL3+, IP-022/Univerza v Ljubljani
- MRIC-UL-IC-BSL3+, IP-022/Univerza v Ljubljani
- MRIC-UL-IC-BSL3+, IP-022/Univerza v Ljubljani
- MRIC-UL-IC-BSL3+, IP-022/Univerza v Ljubljani
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

