Targeting eRNA-Producing Super-Enhancers Regulates TNFα Expression and Mitigates Chronic Inflammation in Mice and Patient-Derived Immune Cells
- PMID: 40692264
- PMCID: PMC12533286
- DOI: 10.1002/advs.202505214
Targeting eRNA-Producing Super-Enhancers Regulates TNFα Expression and Mitigates Chronic Inflammation in Mice and Patient-Derived Immune Cells
Abstract
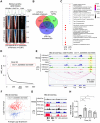
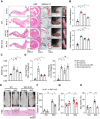
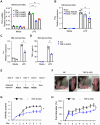
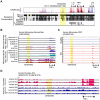
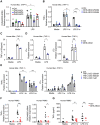
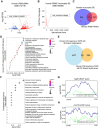
Chronic inflammatory diseases are driven by immune cell dysregulation and overproduction of pro-inflammatory molecules, such as tumor necrosis factor alpha (TNFα). Super-enhancers (SEs) and their enhancer RNAs (eRNAs) are critical gene expression regulators and offer therapeutic potential beyond protein-targeting approaches. This work hypothesizes that targeting eRNAs could reduce chronic inflammation by modulating TNFα expression. This work generates TNF-9 knockout (KO) mice by deleting a Tnfα-regulating enhancer region. These mice exhibit significantly reduced Tnfα levels, improved disease outcomes, and diminished immune cell activation in models of rheumatoid arthritis (RA), psoriasis, and lipopolysaccharide (LPS)-induced sepsis. Integrative epigenomic and transcriptomic analysis identify additional LPS-responsive, eRNA-producing enhancers as therapeutic targets. Antisense oligonucleotide (ASO)-mediated knockdown of TNF-9 eRNA in mouse macrophages demonstrate decreased Tnfα expression and alleviated RA symptoms. Furthermore, ASO-mediated inhibition of the eRNA of the human homolog of TNF-9 similarly reduce TNFα levels. These findings support eRNA-targeted interventions as potential treatment for chronic inflammatory diseases.
Keywords: antisense oligonucleotide; chronic inflammation; enhancer RNA; rheumatoid arthritis; tumor necrosis factor alpha.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
