This is a preprint.
SenSet, a novel human lung senescence cell gene signature, identifies cell-specific senescence mechanisms
- PMID: 40692702
- PMCID: PMC12279080
- DOI: 10.1101/2024.12.21.629928
SenSet, a novel human lung senescence cell gene signature, identifies cell-specific senescence mechanisms
Abstract
Cellular senescence is a major hallmark of aging. Senescence is defined as an irreversible growth arrest observed when cells are exposed to a variety of stressors including DNA damage, oxidative stress, or nutrient deprivation. While senescence is a well-established driver of aging and age-related diseases, it is a highly heterogeneous process with significant variations across organisms, tissues, and cell types. The relatively low abundance of senescence in healthy aged tissues represents a major challenge to studying senescence in a given organ, including the human lung. To overcome this limitation, we developed a Positive-Unlabeled (PU) learning framework to generate a comprehensive senescence marker gene list in human lungs (termed SenSet) using the largest publicly available single-cell lung dataset, the Human Lung Cell Atlas (HLCA). We validated SenSet in a highly complex ex vivo human 3D lung tissue culture model subjected to the senescence inducers bleomycin, doxorubicin, or irradiation, and established its sensitivity and accuracy in characterizing senescence. Using SenSet, we identified and validated cell-type specific senescence signatures in distinct lung cell populations upon aging and environmental exposures. Our study presents the first comprehensive analysis of senescent cells in the healthy aging lung and uncovers cell-specific gene signatures of senescence, presenting fundamental implications for our understanding of major lung diseases, including cancer, fibrosis, chronic obstructive pulmonary disease, or asthma.
Figures
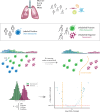





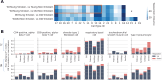
References
-
- Hayflick L. & Moorhead P. S. The serial cultivation of human diploid cell strains. en. Exp. Cell Res. 25, 585–621 (Dec. 1961). - PubMed
-
- Finkel T. & Holbrook N. J. Oxidants, oxidative stress and the biology of ageing. en. Nature 408, 239–247 (Nov. 2000). - PubMed
-
- Campisi J. & d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. en. Nat. Rev. Mol. Cell Biol. 8, 729–740 (Sept. 2007). - PubMed
Publication types
LinkOut - more resources
Full Text Sources
