Identification and validation of DNA methylation-driven gene OSR1 as a novel tumor suppressor for the diagnosis and prognosis of breast cancer
- PMID: 40692711
- PMCID: PMC12277919
- DOI: 10.3389/fgene.2025.1583620
Identification and validation of DNA methylation-driven gene OSR1 as a novel tumor suppressor for the diagnosis and prognosis of breast cancer
Abstract
Introduction: Aberrant DNA methylation plays a critical role in the initiation and progression of cancer, yet its association with breast cancer remains inadequately defined. This study aims to clarify the link between methylation-driven genes and breast cancer pathogenesis.
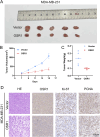
Methods: RNA sequencing and DNA methylation data for breast cancer were retrieved from The Cancer Genome Atlas (TCGA). Prognostically relevant methylation-driven genes were identified by integrating the methylation R package with univariate Cox regression analysis, and OSR1 emerged as the primary candidate. Gene expression profiles and corresponding clinical data were subsequently obtained from TCGA. Differential expression analysis using the Wilcoxon rank-sum test revealed significantly reduced OSR1 expression in breast cancer tissues compared to normal counterparts. Kaplan-Meier survival curves and Cox regression models were applied to assess the prognostic significance of OSR1. Bioinformatic analyses investigated associations between OSR1 expression and clinicopathological features, pathway enrichment, and immune cell infiltration. Experimental validation was conducted by generating OSR1-overexpressing breast cancer cell lines to examine effects on cell viability, migration, and proliferation via phenotypic assays.
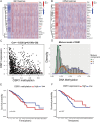
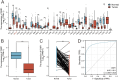
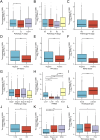
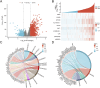
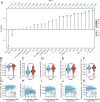
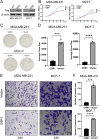
Results: OSR1 expression was significantly reduced in breast cancer tissues and correlated negatively with breast cancer progression. Low OSR1 expression was significantly associated with M stage, HER2 status, PAM50 subtypes, and histological classification, and linked to poorer overall survival outcomes. Functional enrichment implicated OSR1 in pathways related to peptide hormone secretion, peptide transport, metal ion response, and forebrain development. Elevated OSR1 expression was positively correlated with increased infiltration of NK cells, B cells, CD8+ T cells, and dendritic cells. Both in vitro and in vivo studies demonstrated that OSR1 overexpression markedly suppressed breast cancer cell proliferation and migration.
Discussion: These findings confirm OSR1 as a methylation-regulated tumor suppressor gene and underscore its potential as a promising biomarker for individualized therapeutic strategies in breast cancer.
Keywords: DNA methylation; OSR1; TCGA; breast cancer; methylation-driven genes.
Copyright © 2025 Xu, Yang, Cheng, Gan, Huang, Li, Ge, Hu, Ling, Zheng, Zhao, Zhang and Cheng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Dbouk H. A., Weil L. M., Perera G. K., Dellinger M. T., Pearson G., Brekken R. A., et al. (2014). Actions of the protein kinase WNK1 on endothelial cells are differentially mediated by its substrate kinases OSR1 and SPAK. Proc. Natl. Acad. Sci. U. S. A. 111 (45), 15999–16004. 10.1073/pnas.1419057111 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

