Longitudinal profiling of host response and oropharyngeal respiratory microbiome reveals dynamic alterations during recovery from community-acquired pneumonia
- PMID: 40693035
- PMCID: PMC12276579
- DOI: 10.1016/j.bsheal.2025.05.004
Longitudinal profiling of host response and oropharyngeal respiratory microbiome reveals dynamic alterations during recovery from community-acquired pneumonia
Abstract
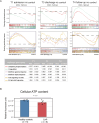
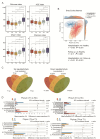
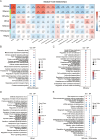
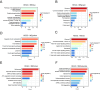
Community-acquired pneumonia (CAP) is a major global health concern, with limited understanding of longitudinal changes in host gene expression and respiratory microbiome throughout disease progression and recovery. To address this gap, we longitudinally collected CAP patients' peripheral blood for transcriptome and oropharyngeal swabs for microbiome analysis from admission to 4 months post infection. Age- and sex-matched volunteers were recruited as controls. We observed CAP patients mounted rapid, effective, and moderate immune responses against infection. Coagulation activation and mitochondrial dysfunction were the striking pathways showing distinct difference in CAP patients compared to controls, and the latter was validated by lower adenosine triphosphate (ATP) levels in the peripheral blood mononuclear cells (PBMCs) of CAP patients. Although transcriptional perturbations gradually decreased, they did not fully recover during the follow-up period. Similarly, persisting oropharyngeal microbiome dysbiosis was observed, characterized by significantly lower alpha diversity and altered taxonomy distribution (P < 0.05). CAP increased the relative abundance of Streptococcus, Veillonella, and Peptostreptococcus, while decreasing that of Haemophilus, Neisseria, and Porphyromonas. Integrated analysis of host response and oropharyngeal microbiome revealed that the relative abundance of Haemophilus, Neisseria, Porphyromonas, and Stomatobaculum were positively related to mitochondrial structure and function pathways, whereas the relative abundance of Prevotella declined over time in patients and positively correlated with anti-pathogen and interferon signaling pathways. These results underscore the persistent impact of CAP on both host immunity and oropharyngeal microbiome, even months after infection, emphasizing the need for long-term follow-up and targeted strategies to facilitate full recovery and restore homeostasis.
Keywords: Community-acquired pneumonia (CAP); Recovery; Respiratory microbiome; Transcriptome.
© 2025 Chinese Medical Association Publishing House. Published by Elsevier B.V.
Figures




References
-
- Lozano R., Naghavi M., Foreman K., Lim S., Shibuya K., Aboyans V., Abraham J., Adair T., Aggarwal R., Ahn S.Y., et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–2128. doi: 10.1016/s0140-6736(12)61728-0. - DOI - PMC - PubMed
-
- Troeger C., Blacker B., Khalil I.A., Rao P.C., Cao J., Zimsen S.R.M., Albertson S.B., Deshpande A., Farag T., Abebe Z., et al. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect. Dis. 2018;18:1191–1210. doi: 10.1016/s1473-3099(18)30310-4. - DOI - PMC - PubMed
-
- Scicluna B.P., Klein Klouwenberg P.M.C., van Vught L.A., Wiewel M.A., Ong D.S., Zwinderman A.H., Franitza M., Toliat M.R., Nürnberg P., Hoogendijk A.J., et al. A Molecular biomarker to diagnose community-acquired pneumonia on intensive care unit admission. Am. J. Respir. Crit. Care Med. 2015;192:826–835. doi: 10.1164/rccm.201502-0355oc. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

