A hypomorphic Mpi mutation unlocks an in vivo tool for studying global N-glycosylation deficiency
- PMID: 40693465
- PMCID: PMC12288971
- DOI: 10.1172/jci.insight.180752
A hypomorphic Mpi mutation unlocks an in vivo tool for studying global N-glycosylation deficiency
Abstract
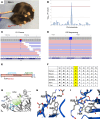
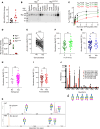
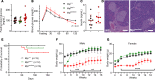
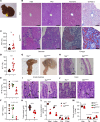
Glycans are one of the 4 major macromolecules essential for life and are the most abundant family of organic molecules. However, in contrast with DNA and RNA, glycan structures have no template; this results in limited tools to study this challenging macromolecule with a diversity of glycan structures. A central bottleneck in studying glycosylation in vivo is that inhibitors and complete KOs are lethal. In a forward genetic screen, we identified a viable, hypomorphic mutation at a conserved site in mannose phosphate isomerase (Mpi) that causes a multisystemic phenotype affecting RBCs, liver, stomach, intestines, skin, size, fat, and fluid balance in mice. The phenotype could be rescued with mannose. Analyses of glycopeptides in mice with this mutation showed a 500% increase in unoccupied N-glycan sites. This is equivalent to a "glycan knockdown," which would be useful for examining the role of glycans in biology and disease. Therefore, we report an in vivo tool to study global N-glycosylation deficiency with tissue-specific targeting and a rescue mechanism with mannose.
Keywords: Gastroenterology; Genetic diseases; Genetics; Glycobiology; Mouse models.
Conflict of interest statement
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

