Dynamic integration and evolutionary trajectory of endogenous IHHNV elements in crustacean genomes
- PMID: 40693514
- PMCID: PMC12287923
- DOI: 10.1093/dnares/dsaf018
Dynamic integration and evolutionary trajectory of endogenous IHHNV elements in crustacean genomes
Abstract
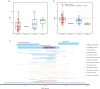
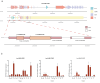
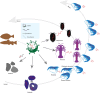
Endogenous viral elements (EVEs) serve as molecular fossils that record the ancient co-evolutionary arms race between viruses and their hosts. In this study, by analyzing 105 host crustacean genomes, we identified 252 infectious hypodermal and haematopoietic necrosis virus-derived EVEs (IHHNV-EVEs), which include 183 ancient and 6 recently inserted EVEs. These IHHNV-EVEs are widely distributed among Decapoda, Thoracica, and Isopoda, with some of them exhibiting a syntenic distribution relative to neighboring host sequences, suggesting that the IHHNV or its ancestor are potential pathogens of these species with a long-time dynamic interaction during the evolutionary history. An expansion of IHHNV-EVEs was observed in decapoda genomes, reflecting a reinforced arm race between decapoda and IHHNV. Notably, we found that nearly all recent IHHNV-EVEs were laboratory contaminants, except for a single authentic integration in Penaeus monodon that persists intact across 16 samples from the 2 populations. These temporal dynamics-ancient genomic stabilization versus modern colonization activity-highlight that EVEs serve as dual archives: historical records of past conflicts and active participants in current evolutionary battles. Our findings redefine viral genomic colonization as a continuum, where ancient EVE fixation coexists with persistent integration processes, providing new insights into host-virus co-evolutionary trajectories.
Keywords: IHHNV-EVE; crustacean; genome; shrimp; virus–host interaction.
© The Author(s) 2025. Published by Oxford University Press on behalf of Kazusa DNA Research Institute.
Conflict of interest statement
None declared.
Figures



References
-
- Katzourakis A, Gifford RJ.. Endogenous viral elements in animal genomes. PLoS Genet. 2010:6:e1001191. https://doi.org/ 10.1371/journal.pgen.1001191 - DOI - PMC - PubMed
-
- Houe V, Bonizzoni M, Failloux AB.. Endogenous non-retroviral elements in genomes of Aedes mosquitoes and vector competence. Emerg Microbes Infect. 2019:8:542–555. https://doi.org/ 10.1080/22221751.2019.1599302 - DOI - PMC - PubMed
-
- Gilbert C, Belliardo C.. The diversity of endogenous viral elements in insects. Curr Opin Insect Sci. 2022:49:48–55. https://doi.org/ 10.1016/j.cois.2021.11.007 - DOI - PubMed
-
- Simmonds P, Aiewsakun P, Katzourakis A.. Prisoners of war – host adaptation and its constraints on virus evolution. Nat Rev Microbiol. 2019:17:321–328. https://doi.org/ 10.1038/s41579-018-0120-2 - DOI - PMC - PubMed
-
- Duggal NK, Emerman M.. Evolutionary conflicts between viruses and restriction factors shape immunity. Nat Rev Immunol. 2012:12:687–695. https://doi.org/ 10.1038/nri3295 - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

