Two strains of Toscana virus show different virulence and replication capacity in mice and cell culture models
- PMID: 40693587
- PMCID: PMC12296115
- DOI: 10.1080/21505594.2025.2535470
Two strains of Toscana virus show different virulence and replication capacity in mice and cell culture models
Abstract
Toscana virus (TOSV), belonging to Phenuiviridae family, is circulating in most Mediterranean countries and is transmitted to humans by infected female sand flies. While most infections are asymptomatic, TOSV is considered as a leading cause of meningitis and encephalitis in humans during summer. Three TOSV genotypes (named A, B, and C) have been identified, although no virus strain belonging to lineage C has been isolated so far. To date, the relationship between TOSV genetic diversity and viral pathogenicity or replication capacity remains unknown. This study aimed to compare two TOSV strains from either lineage A (TOSV-A) or B (TOSV-B) in several cell culture and two mouse models. We showed that TOSV-A replicated more efficiently in BSR and A549 cells, while TOSV-B had a replication advantage in human induced pluripotent stem cells differentiated in neural cells and LL-5 sand fly cells. In vivo, we were unable to detect any virus in the brains of immunocompetent C57BL/6JRj mice infected with either strain of TOSV. On the contrary, we showed that TOSV-A disseminated to the central nervous system of 129/Sv ifnar -/- mice unlike TOSV-B, despite higher viremia of TOSV-B and a greater dissemination of this strain in other organs. The reasons for these differences are not yet known, although we showed that the presence of TOSV neutralizing antibodies in serum was slightly delayed in TOSV-A-infected mice. Altogether, the data presented in this study provide new avenues to study TOSV-induced pathogenesis and ultimately unveil molecular viral determinants modulating TOSV replication capacity.
Keywords: TOSV; Toscana virus; genetic lineage; mouse model; pathogenesis.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
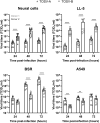

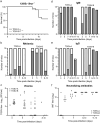
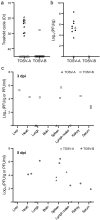
References
-
- Verani P, Ciufolini MG, Nicoletti L, et al. Ecological and epidemiological studies of toscana virus, an arbovirus isolated from phlebotomus. Annal Dell Supc Sanita. 1982;18(3):397–15. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
