Rsp5-mediated ubiquitination of a functional analog of the Rim8 arrestin facilitates Rim pathway activation in Cryptococcus neoformans
- PMID: 40693800
- PMCID: PMC12345238
- DOI: 10.1128/mbio.00732-25
Rsp5-mediated ubiquitination of a functional analog of the Rim8 arrestin facilitates Rim pathway activation in Cryptococcus neoformans
Abstract
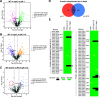
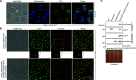
Pathogenic microorganisms use varied cellular processes to adapt to the particular stresses encountered in the infected host. These stresses include rapid alterations in ambient temperature, nutrient availability, and extracellular pH. Fungal pathogens, therefore, rely on the activation of stress response pathways such as the Pal/Rim pathway to adapt to the neutral pH encountered when infecting mammals. While this pathway is conserved among human pathogenic fungi, the proteins required for pH sensing appear to have diverged between different fungal phyla. The opportunistic fungal pathogen Cryptococcus neoformans, a basidiomycete, employs a pH-sensing protein in its Rim pathway that is distinct but functionally analogous to related proteins in well-studied ascomycete fungal systems. We recently characterized protein ubiquitination mediated through the Rsp5 ubiquitin ligase to be required for C. neoformans virulence and for microbial adaptations to host-relevant conditions, including growth at host pH levels. Here, we determined that C. neoformans Rsp5 is specifically required for Rim pathway activation. Using an unbiased screen for proteins that are ubiquitinated by Rsp5 in acidic and alkaline pH, we identified a new component of the C. neoformans Rim pathway that is targeted by Rsp5 for ubiquitination and shares protein features with the ascomycete Rim8/PalF proteins. Rsp5-mediated ubiquitination facilitates protein interactions with Vps23, a downstream trafficking component of the Pal/Rim pathway. Therefore, we define adaptation to ambient pH as one component of the broad cellular roles of Rsp5-mediated ubiquitination, as we explore how protein ubiquitination affects cryptococcal cell physiology, virulence, and microbial interactions with the host.IMPORTANCEExploring the molecular adaptations allowing fungi to grow in an ever-changing environment yields insight into how fungal pathogens adapt to the stresses present in the infected host. The fungal Rim/Pal pathway, activated during alkaline pH stress and during mammalian infection, is of particular interest because of the lack of a homologous pathway in other eukaryotes, providing an opportunity to identify novel targets for antimicrobial therapies with little damage to the host. There is evidence for convergent evolution in this pathway between ascomycetes and basidiomycetes, evident through the functionally converged, but sequence-dissimilar, sensing proteins found in these two fungal groups. Here, we identify the role of ubiquitination in the activation of the Cn Rim pathway. This ubiquitination event is mediated by the Rsp5 E3 Ub ligase and a basidiomycete-specific functional analog of the ascomycete PalF/Rim8 protein that is required for interaction with downstream components of this pathway.
Keywords: Rim101; Rim8; Rra2; Vps23; alkaline pH response pathway; arrestin; fungal pathogenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Gostinčar C, Zajc J, Lenassi M, Plemenitaš A, de Hoog S, Al-Hatmi AMS, Gunde-Cimerman N. 2018. Fungi between extremotolerance and opportunistic pathogenicity on humans. Fungal Divers 93:195–213. doi: 10.1007/s13225-018-0414-8 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

