Characterization of the Nicotinamide Adenine Dinucleotide Biosynthesis Pathway and Regulatory Mechanisms in Streptococcus mutans
- PMID: 40693873
- PMCID: PMC12282500
- DOI: 10.1096/fj.202500944R
Characterization of the Nicotinamide Adenine Dinucleotide Biosynthesis Pathway and Regulatory Mechanisms in Streptococcus mutans
Abstract
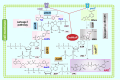
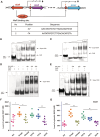
NAD+ and its derivatives, which act as redox coenzymes, are crucial for cellular metabolism and energy production. Nevertheless, the processes by which Streptococcus mutans, a bacterium known for causing dental caries, synthesizes NAD+ are not well elucidated. Through a genome-wide screen, we identified the nicotinic acid salvage pathway and the evolutionarily incomplete PnuC-NadR pathway involved in NAD+ biosynthesis in S. mutans UA159. The nicotinic acid pathway is regulated by SmNiaR, a nicotinic acid-responsive transcription regulator featuring an N-terminal DNA-binding winged helix-turn-helix-like domain and a C-terminal 3-histidine domain. Notably, a single-site amino acid substitution at site K97 in SmNiaR can reverse its DNA-binding ability, an effect mediated by acetylation at this site, which impacts the intracellular production of NAD+ and NADH. Additionally, the deletion of niaR in S. mutans UA159 impaired bacterial proliferation, reduced acid production, and altered biofilm formation, resulting in attenuated virulence in the rat caries model. Conclusively, the regulation of NAD+ homeostasis via SmNiaR contributes significantly to the cariogenic virulence of S. mutans.
Keywords: Streptococcus mutans; NAD+ biosynthesis; acetylation; regulator; virulence.
© 2025 The Author(s). The FASEB Journal published by Wiley Periodicals LLC on behalf of Federation of American Societies for Experimental Biology.
Conflict of interest statement
Integrity Policies: All data presented in this study are accurate and have not been fabricated or manipulated. Proper citation practices have been followed to avoid plagiarism. Permission to Reproduce Material From Other Sources: Request permission: Contact the original publisher or author to request permission. We are unable to provide permission to reproduce this material as it contains confidential information.
The authors declare no conflicts of interest.
Figures


References
-
- Bi J., Wang H., and Xie J., “Comparative Genomics of Nad(p) Biosynthesis and Novel Antibiotic Drug Targets,” Journal of Cellular Physiology 226, no. 2 (2011): 331–340. - PubMed
-
- Chen Y., Ying Y., Lalsiamthara J., et al., “From Bacteria to Biomedicine: Developing Therapies Exploiting NAD+ Metabolism,” Bioorganic Chemistry 142 (2024): 106974. - PubMed
-
- Teramoto H., Inui M., and Yukawa H., “NdnR Is an NAD‐Responsive Transcriptional Repressor of the NdnR Operon Involved in NAD De Novo Biosynthesis in Corynebacterium glutamicum ,” Microbiology 158, no. Pt 4 (2012): 975–982. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

