Identification of a QTL region for tomato brown rugose fruit virus resistance in Solanum pimpinellifolium
- PMID: 40694064
- PMCID: PMC12283848
- DOI: 10.1007/s00122-025-04974-0
Identification of a QTL region for tomato brown rugose fruit virus resistance in Solanum pimpinellifolium
Abstract
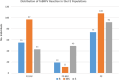
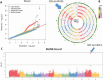
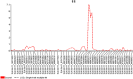
Tomato (Solanum lycopersicum L.), one of the most widely grown vegetables in the world, has been seriously impacted in the past decade by the emerging tomato brown rugose fruit virus (ToBRFV). ToBRFV is a seed-borne tobamovirus, with ability to overcome the commonly used Tm-22 resistance gene in tomato. The objective of this study was to conduct quantitative trait locus (QTL) mapping and identify single-nucleotide polymorphism (SNP) markers associated with ToBRFV resistance in tomato. Two F2 populations were used for QTL mapping: One derived from a cross between S. pimpinellifolium USVL333 (PI 390718) × USVL332 (PI 390717) and another from 'Moneymaker' × USVL332 (PI 390717), with population sizes of 195 and 79 plants, respectively. The resistance trait was derived from the S. pimpinellifolium accession USVL332 (PI 390717). A major QTL for ToBRFV resistance was identified on chromosome 11 (SL4.0ch11), with the peak located at approximately 46.84 Mbp. This QTL spans a 22-kb interval between 46,825,788 bp and 46,847,421 bp, as determined through both genome-wide association study (GWAS) and QTL linkage mapping. Three SNP markers, SL4.0ch11_46825788, SL4.0ch11_46847421, and SL4.0ch11_46850215, demonstrated the most significant association with high LOD values (LOD = 13 in the Blink model) in GWAS analysis. In this genomic region, two disease resistance gene analogs, Solyc11g062150 (TIR-NBS-LRR resistance protein, Toll-Interleukin receptor) and Solyc11g062180 (disease resistance protein, leucine-rich repeat), were identified, which may serve as candidates for ToBRFV resistance. The QTL identified in this study could be valuable for plant breeders in facilitating tomato breeding with ToBRFV resistance.
© 2025. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Declarations. Conflict of interest: There is no conflict of interest in this research.
Figures


References
-
- Alexander LJ (1963) Transfer of a dominant type of resistance to the four known Ohio pathogenic strains of tobacco mosaic virus (TMV) from Lycopersiconperuvianum to L. esculentum. Phytopathology 53:869
-
- Ashkenazi V, Rotem Y, Ecker R, Nashilevitz S, Barom N (2018) Tolerance in plants of Solanum lycopersicum to the tobamovirus tomato brown rugose fruit virus (TBRFV). WO Patent 2018/219941. https://patentscope.wipo.int/search/es/detail.jsf?docId=WO2018219941
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

