OGG1 augments the transcriptional activation of Foxp3 to promote iTreg differentiation for IBD alleviation
- PMID: 40694333
- PMCID: PMC12318175
- DOI: 10.1073/pnas.2424733122
OGG1 augments the transcriptional activation of Foxp3 to promote iTreg differentiation for IBD alleviation
Abstract
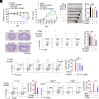
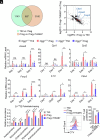
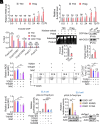
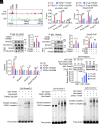
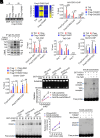
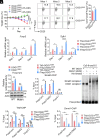
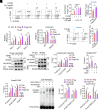
8-oxo-7,8-dihydroguanine (8-oxoG), the most frequent form of oxidative-DNA-base lesion caused by ROS, is recognized and repaired by 8-oxoguanine DNA glycosylase 1 (OGG1) through base excision repair (BER) pathway. Beyond its role in DNA repair, OGG1 has been shown to promote transcriptional activation of proinflammatory mediators and contribute to both acute and chronic lung inflammation. However, pioneering studies have shown an anti-inflammation role for OGG1 in inflammatory bowel disease (IBD), but its underlying molecular mechanism remains unclear. In the present study, we unveiled that OGG1 plays an important role in the differentiation of inducible regulatory T cells (iTregs). Binding of OGG1 to 8-oxoG facilitated the recruitment of Smad3 to the Foxp3 promoter, leading to the transcriptional activation. Moreover, OGG1 binding promoted demethylation of CpG sites in the conserved noncoding sequence 2 (CNS2) region of Foxp3 by decreasing Dnmt1 occupancy and enhancing recruitment of Tet1/2. Notably, the S326C variant-a naturally occurring polymorphism in humans-was more effective than the wild-type protein in promoting iTreg differentiation and showed a negative correlation with IBD incidence. Furthermore, treatment with O8, a selective OGG1 inhibitor that blocks base excision activity without affecting substrate binding, significantly alleviated IBD in a mouse model, suggesting a promising therapeutic strategy. Together, these findings extend the understanding of OGG1's epigenetic role in transcriptional regulation and highlight its protective function in inflammatory diseases, potentially shaped by aerobic evolution.
Keywords: Foxp3; IBD; OGG1; Smad3; iTreg differentiation.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures

Similar articles
-
Pollen-induced oxidative DNA damage response regulates miRNAs controlling allergic inflammation.Am J Physiol Lung Cell Mol Physiol. 2017 Dec 1;313(6):L1058-L1068. doi: 10.1152/ajplung.00141.2017. Epub 2017 Aug 10. Am J Physiol Lung Cell Mol Physiol. 2017. PMID: 28798252 Free PMC article.
-
The BCL11A transcription factor stimulates the enzymatic activities of the OGG1 DNA glycosylase.Biol Chem. 2024 Sep 16;405(11-12):711-726. doi: 10.1515/hsz-2024-0088. Print 2024 Dec 17. Biol Chem. 2024. PMID: 39272221 Free PMC article.
-
The Relationship Between CSF1R Signaling, Monocyte-Macrophage Differentiation, and Susceptibility to Inflammatory Bowel Disease.Cell Mol Gastroenterol Hepatol. 2025;19(8):101510. doi: 10.1016/j.jcmgh.2025.101510. Epub 2025 Mar 26. Cell Mol Gastroenterol Hepatol. 2025. PMID: 40154882 Free PMC article. Review.
-
Genome-wide CRISPR screen in human T cells reveals regulators of FOXP3.Nature. 2025 Jun;642(8066):191-200. doi: 10.1038/s41586-025-08795-5. Epub 2025 Mar 26. Nature. 2025. PMID: 40140585
-
Management of urinary stones by experts in stone disease (ESD 2025).Arch Ital Urol Androl. 2025 Jun 30;97(2):14085. doi: 10.4081/aiua.2025.14085. Epub 2025 Jun 30. Arch Ital Urol Androl. 2025. PMID: 40583613 Review.
References
-
- Kasai H., et al. , Formation of 8-hydroxyguanine moiety in cellular DNA by agents producing oxygen radicals and evidence for its repair. Carcinogenesis 7, 1849–1851 (1986). - PubMed
-
- Neeley W. L., Essigmann J. M., Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem. Res. Toxicol. 19, 491–505 (2006). - PubMed
MeSH terms
Substances
Grants and funding
- P01 AI062885/AI/NIAID NIH HHS/United States
- QT202304/Department of Science and Technology of Jilin Province ()
- JJKH20250323KJ/Education Department of Jilin Province ()
- 2025YFE0107900/Key Special Project for International Science and Technology Innovation Cooperation between Governments under the National Key R&D program
- JJKH20231314KJ/Education Department of Jilin Province ()
LinkOut - more resources
Full Text Sources
Research Materials

