Marek's disease virus replication in chicken skin reconstructed in vitro: evidence for viral particles in corneocytes
- PMID: 40694389
- PMCID: PMC12283110
- DOI: 10.1099/jgv.0.002123
Marek's disease virus replication in chicken skin reconstructed in vitro: evidence for viral particles in corneocytes
Abstract
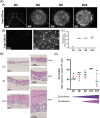
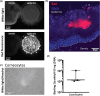
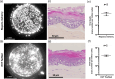
Marek's disease (MD) is a lethal lymphoma of chickens, which is caused by MD virus (MDV), an alphaherpesvirus. MDV infects epithelial cells of the skin appendages, notably feather follicles, replicates in these cells and is shed into the environment exclusively from these tissues. Here, we tested whether chicken skin equivalents (SEs) can be used to model MDV infection. Primary chicken keratinocytes were seeded on a suspension of fibroblasts in collagen and induced to terminally differentiate at the air-liquid interface. A recombinant MDV expressing the Katushka fluorescent protein (MDV-KAT) was introduced into SEs by seeding primary keratinocytes together with MDV-KAT-infected keratinocytes of the K8 cell line. KAT-mediated fluorescence increased during the culture of infected SEs, indicating virus infection and replication, while the expression of keratinocyte differentiation markers was not significantly altered by MDV infection. MDV did not spread to the dermal compartment of SEs but localized to the upper layers of the epidermis. Viral particles were readily observed by electron microscopy in living keratinocytes and for the first time in cornified keratinocytes of the outermost layer of infected SEs, suggesting that viral elements can be released into the environment. Finally, we demonstrated that two fluorescent vaccine strains of MDV, Rispens and herpesvirus of turkey, can infect and replicate in SEs. Taken together, this study establishes chicken SEs as an in vitro model for essential steps of MDV infection.
Keywords: 3D in vitro models; Marek’s disease virus; chicken; herpesvirus; keratinocyte; viral particles.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures





Similar articles
-
Amino acid insertion in the Meq protein of Marek's disease virus, an avian oncogenic herpesvirus, accelerates tumorigenesis.Microbiol Spectr. 2025 Aug 5;13(8):e0336824. doi: 10.1128/spectrum.03368-24. Epub 2025 Jul 17. Microbiol Spectr. 2025. PMID: 40673706 Free PMC article.
-
The Meq Genes of Nigerian Marek's Disease Virus (MDV) Field Isolates Contain Mutations Common to Both European and US High Virulence Strains.Viruses. 2024 Dec 31;17(1):56. doi: 10.3390/v17010056. Viruses. 2024. PMID: 39861844 Free PMC article.
-
Marek's Disease Virus (MDV) Meq Oncoprotein Plays Distinct Roles in Tumor Incidence, Distribution, and Size.Viruses. 2025 Feb 14;17(2):259. doi: 10.3390/v17020259. Viruses. 2025. PMID: 40007015 Free PMC article.
-
Antiretrovirals for reducing the risk of mother-to-child transmission of HIV infection.Cochrane Database Syst Rev. 2011 Jul 6;(7):CD003510. doi: 10.1002/14651858.CD003510.pub3. Cochrane Database Syst Rev. 2011. PMID: 21735394
-
Immunogenicity and seroefficacy of pneumococcal conjugate vaccines: a systematic review and network meta-analysis.Health Technol Assess. 2024 Jul;28(34):1-109. doi: 10.3310/YWHA3079. Health Technol Assess. 2024. PMID: 39046101 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

