Integrating pulmonary and systemic transcriptomes to characterize lung injury after pediatric hematopoietic stem cell transplant
- PMID: 40694427
- PMCID: PMC12487673
- DOI: 10.1172/jci.insight.194440
Integrating pulmonary and systemic transcriptomes to characterize lung injury after pediatric hematopoietic stem cell transplant
Abstract
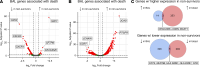
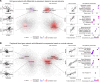
Hematopoietic stem cell transplantation (HCT) is a potentially life-saving therapy but can lead to lung injury due to chemoradiation toxicity, infection, and immune dysregulationWe previously showed that bronchoalveolar lavage (BAL) transcriptomes representing pulmonary inflammation and cellular injury can phenotype post-HCT lung injury and predict mortality. To test whether peripheral blood might be a suitable surrogate for BAL, we compared 210 paired BAL and blood transcriptomes obtained from 166 pediatric patients with HCT at 27 hospitals. BAL and blood RNA abundance showed minimal correlation at the level of individual genes, gene set enrichment scores, imputed cell fractions, and T and B cell receptor clonotypes. Instead, we identified significant site-specific transcriptional programs. In BAL, pathways related to immunity, hypoxia, and epithelial mesenchymal transition were tightly coexpressed and linked to mortality. In contrast, in blood, expression of endothelial injury, DNA repair, and cellular metabolism pathways was associated with mortality. Integration of paired BAL and blood transcriptomes dichotomized patients into 2 groups with significantly different rates of hypoxia and clinical outcomes within 1 week of BAL. These findings reveal a compartmentalized injury response, where BAL and blood transcriptomes provide distinct but complementary insights into local and systemic mechanisms of post-HCT lung injury.
Keywords: Bone marrow transplantation; Immunology; Inflammation; Pulmonology; Stem cell transplantation; Transcriptomics.
Conflict of interest statement
Figures




Update of
-
Integrating Pulmonary and Systemic Transcriptomic Profiles to Characterize Lung Injury after Pediatric Hematopoietic Stem Cell Transplant.medRxiv [Preprint]. 2025 Apr 1:2025.03.31.25324969. doi: 10.1101/2025.03.31.25324969. medRxiv. 2025. Update in: JCI Insight. 2025 Jul 22;10(17):e194440. doi: 10.1172/jci.insight.194440. PMID: 40236411 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

