Different SARS-CoV-2 variants inhibited by RRM designed peptide
- PMID: 40694559
- PMCID: PMC12282855
- DOI: 10.1371/journal.pone.0327582
Different SARS-CoV-2 variants inhibited by RRM designed peptide
Abstract
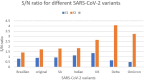
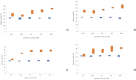
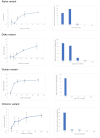
The SARS-CoV-2 virus has mutated over time, resulting in variations of circulating viral variants, which may impact the virus's properties, such as transmission or the severity of symptoms. Thus, there is a need for comprehensive approach less dependent of viral variants. For that purpose, we have employed the innovative and unique Resonant Recognition Model (RRM) to identify the common characteristics of all variants of SARS-CoV-2 virus and based on these characteristics, we have explored the possibility to develop approach against SARS-CoV-2 infection, less dependent on viral variants. This paper is a continuation of our previous research, where we have used the RRM model to design de novo peptide CovA, capable to prevent SARS-CoV-2 interaction with ACE2 receptor on host cells. Using Inhibitor Screening Assay Kits and viral replication in SARS-CoV-2/Vero E6 cells model, it has been previously shown that CovA can prevent viral interaction with receptor and viral replication in host cells. This test has been done on Wuhan variant only. Here, we have tested this RRM designed peptide CovA for efficiency with some other different SARS-CoV-2 variants. It has been shown here, that CovA act on all tested viral variants, but with different efficiency. Apart from representing the basis of new COVID-19 drugs discovery, this research once again presents the ability of RRM model to design de novo bioactive peptides with desired biological function.
Copyright: © 2025 Krapez et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
No competing interests.
Figures
References
-
- Kupferschmidt K. New coronavirus variants could cause more reinfections, require updated vaccines. Science. 2021. doi: 10.1126/science.abg6028 - DOI
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

