FEN1 is critical for rapid single-strand break repair in G1 phase
- PMID: 40694846
- PMCID: PMC12282947
- DOI: 10.1093/nar/gkaf710
FEN1 is critical for rapid single-strand break repair in G1 phase
Abstract
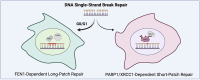
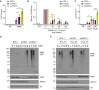
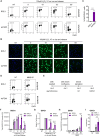
Flap endonuclease 1 (FEN1)-dependent long-patch repair has been considered a minor sub-pathway of DNA single-strand break repair (SSBR), activated only when short-patch repair is not feasible. However, the significance of long-patch repair in living cells remains unclear. Here, we employed human RPE-1 cells with FEN1 deletion to compare the requirements for short- and long-patch pathways for the rapid repair of various types of DNA single-strand breaks (SSBs). We found that SSBs arising from abortive topoisomerase 1 activity are repaired efficiently without FEN1. In contrast, the rapid repair of SSBs arising during base excision repair following treatment with methyl methanesulphonate (MMS) or following treatment with hydrogen peroxide (H2O2) exhibits an unexpectedly high dependence on FEN1. Indeed, in G1 phase, FEN1 deletion slows the rate of SSBR to a similar or even greater extent than deletion of the short-patch repair proteins XRCC1 or POLβ. As expected, the combined deletion of FEN1 with XRCC1 or POLβ has an additive or synergistic effect, severely attenuating SSBR rates after MMS or H2O2 exposure. These data highlight an unanticipated requirement for FEN1 in the rapid repair of SSBs in human cells, challenging the prevailing view that long-patch repair is a minor sub-pathway of SSBR.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

