Mouse metastable epialleles are extremely rare
- PMID: 40694849
- PMCID: PMC12282951
- DOI: 10.1093/nar/gkaf624
Mouse metastable epialleles are extremely rare
Abstract
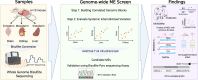
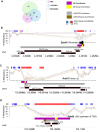
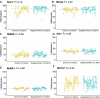
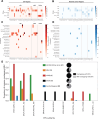
Metastable epialleles (MEs) are genomic loci at which epigenetic marks are established stochastically during early embryonic development and maintained during subsequent differentiation and throughout life, leading to stable epigenetic and phenotypic variation among genetically identical individuals. Although MEs were first described in mice over 20 years ago, the extent of epigenetic metastability in the mouse genome remains unknown. We present the first unbiased genome-wide screen for MEs in mice. Using deep whole-genome bisulfite sequencing across tissues derived from the three embryonic germ layers in isogenic C57BL/6J mice, we identified only 29 MEs, precisely localizing them and documenting their rarity. Consistent with recent findings, we found no effects of maternal dietary methyl donor supplementation on ME methylation in the offspring, challenging previous assertions that MEs generally exhibit developmental plasticity. Most but not all MEs are associated with intracisternal A-particle (IAP) elements, tending to localize to the 5' end of the IAP. Additionally, we discovered autosomal regions at which systemic interindividual variation in DNA methylation is associated with sex, providing insights into sex-associated epigenetic development that apparently precedes sexual differentiation. Our findings indicate that expression of transcription factors, including CCCTC-binding factor (CTCF) and specific KRAB zinc finger proteins during early embryonic development, plays a key role in orchestrating stochastic establishment and/or maintenance of DNA methylation at metastable transposable elements. Overall, these findings advance our understanding of the genomic determinants of epigenetic metastability and suggest that interindividual epigenetic variation at MEs is unlikely to be a major determinant of phenotypic variation among isogenic mice.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

