Understanding the heterogeneity of pancreatic ductal adenocarcinoma
- PMID: 40694876
- PMCID: PMC12304713
- DOI: 10.1016/j.tranon.2025.102479
Understanding the heterogeneity of pancreatic ductal adenocarcinoma
Abstract
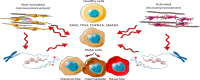
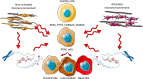
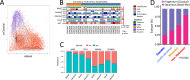
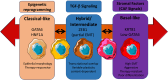
Pancreatic ductal adenocarcinoma (PDAC) is a highly aggressive, treatment-resistant cancer characterized by extensive inter- and intra-tumoral heterogeneity. Although over 95 % of cases harbor KRAS mutations and commonly altered tumor suppressors like TP53, SMAD4, and CDKN2A, these genetic changes alone do not fully explain PDAC variability. We propose a paradigm shift: PDAC heterogeneity is not solely genetic but also shaped by epigenetic regulation and the tumor microenvironment. Traditional transcriptomic classifications define PDAC into fixed subtypes, primarily classical and basal-like, but we argue these are not static categories. Instead, PDAC phenotypes exist along a dynamic continuum influenced by stromal interactions and epigenetic cues. This model challenges the binary classification view. We show that transitions from classical to basal-like states are gradual and reversible, driven by tumor-stroma crosstalk and chromatin remodeling. Such plasticity underpins tumor adaptation, resistance, and progression. Embracing this dynamic framework offers novel therapeutic opportunities.
Keywords: Epigenetics; Metastases; PDAC; Tumor heterogeneity; Tumor microenvironment; Tumor progression.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of competing interest The authors disclose no conflicts of interest.
Figures
References
-
- Nicolle R., Blum Y., Marisa L., Loncle C., Gayet O., Moutardier V., Turrini O., Giovannini M., Bian B., Bigonnet M., et al. Pancreatic adenocarcinoma therapeutic targets revealed by tumor-stroma cross-talk analyses in patient-derived xenografts. Cell Rep. 2017;21:2458–2470. doi: 10.1016/j.celrep.2017.11.003. - DOI - PMC - PubMed
-
- Moffitt R.A., Marayati R., Flate E.L., Volmar K.E., Loeza S.G.H., Hoadley K.A., Rashid N.U., Williams L.A., Eaton S.C., Chung A.H., et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 2015;47:1168–1178. doi: 10.1038/ng.3398. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

