Testing for the Genomic Footprint of Conflict Between Life Stages in an Angiosperm and Moss Species
- PMID: 40695727
- PMCID: PMC12351284
- DOI: 10.1093/gbe/evaf138
Testing for the Genomic Footprint of Conflict Between Life Stages in an Angiosperm and Moss Species
Abstract
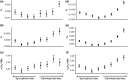
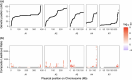
The maintenance of genetic variation by balancing selection is of considerable interest to evolutionary biologists. An important but understudied potential driver of balancing selection is antagonistic pleiotropy between diploid and haploid stages of the plant life cycle. Despite sharing a common genome, sporophytes (2n) and gametophytes (n) may undergo differential or even opposing selection. Theoretical work suggests antagonistic pleiotropy between life stages can generate balancing selection and maintain genetic variation. Despite the potential for far-reaching consequences of gametophytic selection, empirical tests of its pleiotropic effects (neutral, synergistic, or antagonistic) on sporophytes are generally lacking. Here, we examined the population genomic signals of selection across life stages in the angiosperm Rumex hastatulus and the moss Ceratodon purpureus. We compared gene expression between life stages and sexes, combined with neutral diversity statistics and the analysis of the distribution of fitness effects. In contrast to what would be predicted under balancing selection due to antagonistic pleiotropy, we found that unbiased genes between life stages were under stronger purifying selection, likely explained by a predominance of synergistic pleiotropy between life stages and strong purifying selection on broadly expressed genes. In addition, we found that 30% of candidate genes under balancing selection in R. hastatulus were located within inversion polymorphisms. Our findings provide novel insights into the genome-wide characteristics and consequences of plant gametophytic selection.
Keywords: antagonistic pleiotropy; balancing selection; gametophytic selection; gene expression; intralocus conflict.
© The Author(s) 2025. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures



References
-
- Alexa A, Rahnenfuhrer J. topGO: Enrichment analysis for gene ontology [Internet]. 2022. [accessed 2022 May 20]. Available from: https://bioconductor.org/packages/topGO/
-
- Andrews S. FastQC: a quality control tool for high throughput sequence data [Internet]. 2010. [accessed 2017 Dec 21]. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995:57:289–300. 10.1111/j.2517-6161.1995.tb02031.x. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

