Aberrant EZHIP expression drives tumorigenesis in osteosarcoma
- PMID: 40695784
- PMCID: PMC12284272
- DOI: 10.1038/s41467-025-61558-8
Aberrant EZHIP expression drives tumorigenesis in osteosarcoma
Abstract
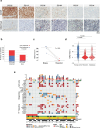
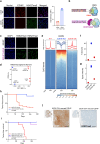
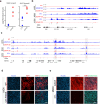
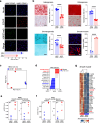
Osteosarcomas (OS) are aggressive bone tumors known for their extensive structural variations and rare recurrent oncogenic driver mutations. In this study, we identify ectopic expression of the oncohistone-mimic EZHIP in 20% of patients across two independent OS cohorts. We demonstrate that reduced deposition of the repressive H3K27me3 mark correlates with poor histological response to neoadjuvant therapy, serving as a predictor of patient outcomes. Through gain- and loss-of-function experiments, we provide functional evidence of the oncogenic activity of EZHIP in enhancing the aggressive characteristics of OS both in vitro and in vivo. EZHIP-induced epigenetic reprogramming reactivates developmental pathways and impedes the differentiation of mesenchymal progenitors, pushing them towards smooth muscle lineage commitment at the cost of other fates. Targeting residual H3K27me3 with EZH2 inhibitors may offer therapeutic benefit in EZHIP-expressing OS. Our findings highlight EZHIP expression as a prevalent driver in OS, offering insights into its pathogenic mechanisms and potential therapeutic strategies for this debilitating cancer.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no potential conflicts of interest.
Figures


References
-
- Grobner, S. N. et al. The landscape of genomic alterations across childhood cancers. Nature555, 321–327 (2018). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

