MARCO expression on myeloid-derived suppressor cells is essential for their differentiation and immunosuppression
- PMID: 40695812
- PMCID: PMC12283920
- DOI: 10.1038/s41420-025-02627-1
MARCO expression on myeloid-derived suppressor cells is essential for their differentiation and immunosuppression
Abstract
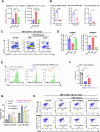
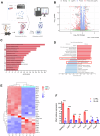
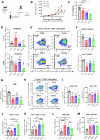
Myeloid-derived suppressor cells (MDSCs) significantly contribute to the immunosuppressive tumor microenvironment (TME), and targeted inhibition of MDSCs is a potential therapeutic strategy against cancer. Here, we identify macrophage receptor with collagenous structure (MARCO) as a critical regulator of MDSC differentiation and immunosuppression in breast cancer. The present study demonstrates that MARCO is expressed on MDSCs, and breast tumor-derived exosomes (TDEs) enriched with macrophage migration inhibitory factor (MIF) promote MDSC differentiation and amplify immunosuppressive activity by up-regulating MARCO. Genetic ablation of MARCO in a murine breast cancer model attenuated tumor growth, accompanied by reduced monocytic MDSCs (M-MDSCs) and total tumor-associated macrophages (TAMs), along with enhanced infiltration of CD8+ T cells and natural killer (NK) cells. Furthermore, we developed a specific MARCO down-regulation-promoting monoclonal antibody that impeded TDE-induced MDSC differentiation and immunosuppression. In vivo, MARCO down-regulating antibody suppressed tumor growth and reprogrammed the TME by diminishing immunosuppressive MDSCs and TAMs and revitalizing CD8+ T cells and NK cells. Strikingly, combining the MARCO down-regulating antibody with PD-1 blockade synergistically enhanced anti-tumor efficacy. This work establishes MARCO as a key regulator of MDSC-mediated immunosuppression and presents a compelling case for the inclusion of MARCO as a therapeutic target in cancer immunotherapy.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
PMN-MDSCs are responsible for immune suppression in anti-PD-1 treated TAP1 defective melanoma.Clin Transl Oncol. 2025 Jul;27(7):3073-3083. doi: 10.1007/s12094-024-03840-7. Epub 2025 Jan 18. Clin Transl Oncol. 2025. PMID: 39825997
-
The co-location of MARCO+ tumor-associated macrophages and CTSE+ tumor cells determined the poor prognosis in intrahepatic cholangiocarcinoma.Hepatology. 2025 Jul 1;82(1):25-41. doi: 10.1097/HEP.0000000000001138. Epub 2024 Oct 29. Hepatology. 2025. PMID: 39471066 Free PMC article.
-
Deletion of gasdermin D promotes granulocytic myeloid-derived suppressor cell differentiation by decreased release of mitochondrial DNA to promote tumor escape.Cancer Immunol Immunother. 2025 Jul 25;74(9):277. doi: 10.1007/s00262-025-04104-1. Cancer Immunol Immunother. 2025. PMID: 40711527 Free PMC article.
-
Systemic treatments for metastatic cutaneous melanoma.Cochrane Database Syst Rev. 2018 Feb 6;2(2):CD011123. doi: 10.1002/14651858.CD011123.pub2. Cochrane Database Syst Rev. 2018. PMID: 29405038 Free PMC article.
-
Immunosuppressive T-cell antibody induction for heart transplant recipients.Cochrane Database Syst Rev. 2013 Dec 2;2013(12):CD008842. doi: 10.1002/14651858.CD008842.pub2. Cochrane Database Syst Rev. 2013. PMID: 24297433 Free PMC article.
References
-
- Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3:453–8. 10.1038/nrc1098. - PubMed
-
- de Visser KE, Joyce JA. The evolving tumor microenvironment: from cancer initiation to metastatic outgrowth. Cancer Cell. 2023;41:374–403. 10.1016/j.ccell.2023.02.016. - PubMed
-
- Almand B, Clark JI, Nikitina E, van Beynen J, English NR, Knight SC, et al. Increased production of immature myeloid cells in cancer patients: a mechanism of immunosuppression in cancer. J Immunol. 2001;166:678–89. 10.4049/jimmunol.166.1.678. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

