A quantum machine learning framework for predicting drug sensitivity in multiple myeloma using proteomic data
- PMID: 40695887
- PMCID: PMC12284003
- DOI: 10.1038/s41598-025-06544-2
A quantum machine learning framework for predicting drug sensitivity in multiple myeloma using proteomic data
Abstract
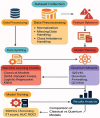
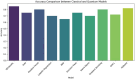
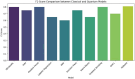
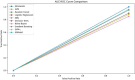
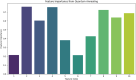
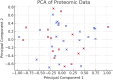
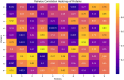
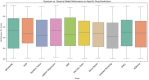
In this paper, we introduce QProteoML, a new quantum machine learning (QML) framework for predicting drug sensitivity in Multiple Myeloma (MM) using high-dimensional proteomic data. MM, an extremely heterogeneous condition, displays often mixed responses to treatment, with a large number of patients showing drug resistance to proteasome inhibitors and immune modulatory agents. However, the methods previously used for genomic and proteomic data analysis techniques are plagued by issues of high dimensionality, imbalanced class distribution and feature redundancy, which work against the accurate predictability and generalizability of such methods. These are compounded by the so-called "curse of dimensionality", with dimensions far outnumbering samples, hence classical model overfitting. In this work, we present QProteoML as an integration of quantum techniques purposefully developed to deal with high-dimensional, imbalanced and redundant data. The framework integrates a combination of Quantum Support Vector Machine (QSVM), Quantum Principal Component Analysis (qPCA), Quantum Annealing (QA) for feature selection and Quantum Generative Adversarial Networks (QGANs) for data augmentation. These quantum algorithms exploit certain quantum phenomena (superposition and entanglement) to perform modelling of nonlinear relationships, dimensionality reduction, and class-imbalance issues. QSVM employs quantum kernels to map data into a higher-dimensional Hilbert space, so that the model can detect complex patterns in MM drug resistance. qPCA reduces dimensionality without loss of important variance, and thus improves computation efficiency. In addition, Quantum Annealing successfully extracts the most informative biomarkers with low redundancy. QProteoML was experimentally tested by comparing accuracy, F1 score and AUC ROC between classical machine learning models such as Support Vector Machine (SVM), Random Forest (RF), Logistic Regression (LR), and K-Nearest Neighbors (KNN). Our results demonstrate that QProteoML performs better than classical models, particularly in identifying the drug resistant minority class of patients. Additionally, the model is interpretable and stresses important biomarkers of drug sensitivity in MM. This research opens the possibility of quantum machine learning in personalised medicine for Multiple Myeloma. It demonstrates that quantum algorithms can perform complex biological data suggesting more reliable and accurate drug sensitivity predictions. Future research will be directed toward clinical validation of the given system with larger and more diverse cohorts of MM patients; the integration of quantum hardware for practical applications.
Keywords: Biomarker discovery; Drug sensitivity prediction; Multiple myeloma; Proteomics data; QSVM; Quantum machine learning (QML).
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures
Similar articles
-
Are Current Survival Prediction Tools Useful When Treating Subsequent Skeletal-related Events From Bone Metastases?Clin Orthop Relat Res. 2024 Sep 1;482(9):1710-1721. doi: 10.1097/CORR.0000000000003030. Epub 2024 Mar 22. Clin Orthop Relat Res. 2024. PMID: 38517402
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Stabilizing machine learning for reproducible and explainable results: A novel validation approach to subject-specific insights.Comput Methods Programs Biomed. 2025 Sep;269:108899. doi: 10.1016/j.cmpb.2025.108899. Epub 2025 Jun 21. Comput Methods Programs Biomed. 2025. PMID: 40570739
-
Bisphosphonates in multiple myeloma: an updated network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 18;12(12):CD003188. doi: 10.1002/14651858.CD003188.pub4. Cochrane Database Syst Rev. 2017. PMID: 29253322 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
References
-
- Mosquera Orgueira, A. et al. Survival prediction and treatment optimization of multiple myeloma patients using machine-learning models based on clinical and gene expression data. Leukemia35(10), 2924–2935 (2021). - PubMed
-
- Guerrero, C. et al. A machine learning model based on tumor and immune biomarkers to predict undetectable MRD and survival outcomes in multiple myeloma. Clin. Cancer Res.28(12), 2598–2609 (2022). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

